import numpy as np
import intake
import matplotlib.pyplot as plt
import hvplot.xarray # noqa: F401
import holoviews as hvIn this notebook, we aim to demonstrate how C-band (4–8 GHz, wavelengths of approximately 3.75–7.5 cm) and L-band (1–2 GHz, wavelengths of approximately 15–30 cm) radio frequencies differ for different land covers and times of the year. In addition, we’ll look at co- and cross-polarized backscattering:
- Sentinel-1 (C-band)
- VV
- VH
- Alos-2 (L-band):
- HH
- HV
5.1 Data Loading
We load the data again with the help of intake.
url = "https://huggingface.co/datasets/martinschobben/microwave-remote-sensing/resolve/main/microwave-remote-sensing.yml"
cat = intake.open_catalog(url)
fused_ds = cat.fused.read()
fused_ds/home/runner/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/intake/readers/readers.py:1327: UserWarning: The specified chunks separate the stored chunks along dimension "time" starting at index 1. This could degrade performance. Instead, consider rechunking after loading.
return open_dataset(data.url, **kw)<xarray.Dataset> Size: 460MB
Dimensions: (time: 5, y: 1528, x: 2508, sensor: 4)
Coordinates:
crs int64 8B ...
* sensor (sensor) object 32B 's1_VH' 's1_VV' 'alos_HV' 'alos_HH'
spatial_ref int64 8B ...
* time (time) datetime64[ns] 40B 2022-06-30 2022-07-31 ... 2022-10-31
* x (x) float64 20kB 4.769e+06 4.769e+06 ... 4.794e+06 4.794e+06
* y (y) float64 12kB 1.397e+06 1.397e+06 ... 1.382e+06 1.382e+06
Data variables:
LAI (time, y, x) float64 153MB dask.array<chunksize=(1, 1528, 2508), meta=np.ndarray>
gam0 (time, sensor, y, x) float32 307MB dask.array<chunksize=(1, 4, 1528, 2508), meta=np.ndarray>The loaded data contains the Leaf Area Index (LAI), which is used as an estimate of foliage cover of forest canopies. So high LAI is interpreted as forested area, whereas low values account for less vegetated areas (shrubs, grass-land, and crops).
First we’ll have a look at the mean and standard deviation of LAI over all timeslices. This can be achieved by using the mean and std methods of the xarray object and by supplying a dimension over which these aggregating operations will be applied. We use the dimension “time”, thereby flattening the cube to a 2-D array with dimensions x and y.
fig, ax = plt.subplots(1, 2, figsize=(15, 6))
LAI_dc = fused_ds.LAI
LAI_mean = LAI_dc.mean("time")
LAI_std = LAI_dc.std("time")
LAI_mean.plot(ax=ax[0], vmin=0, vmax=6).axes.set_aspect("equal")
LAI_std.plot(ax=ax[1], vmin=0, vmax=3).axes.set_aspect("equal")
plt.tight_layout()/home/runner/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/dask/array/numpy_compat.py:57: RuntimeWarning: invalid value encountered in divide
x = np.divide(x1, x2, out)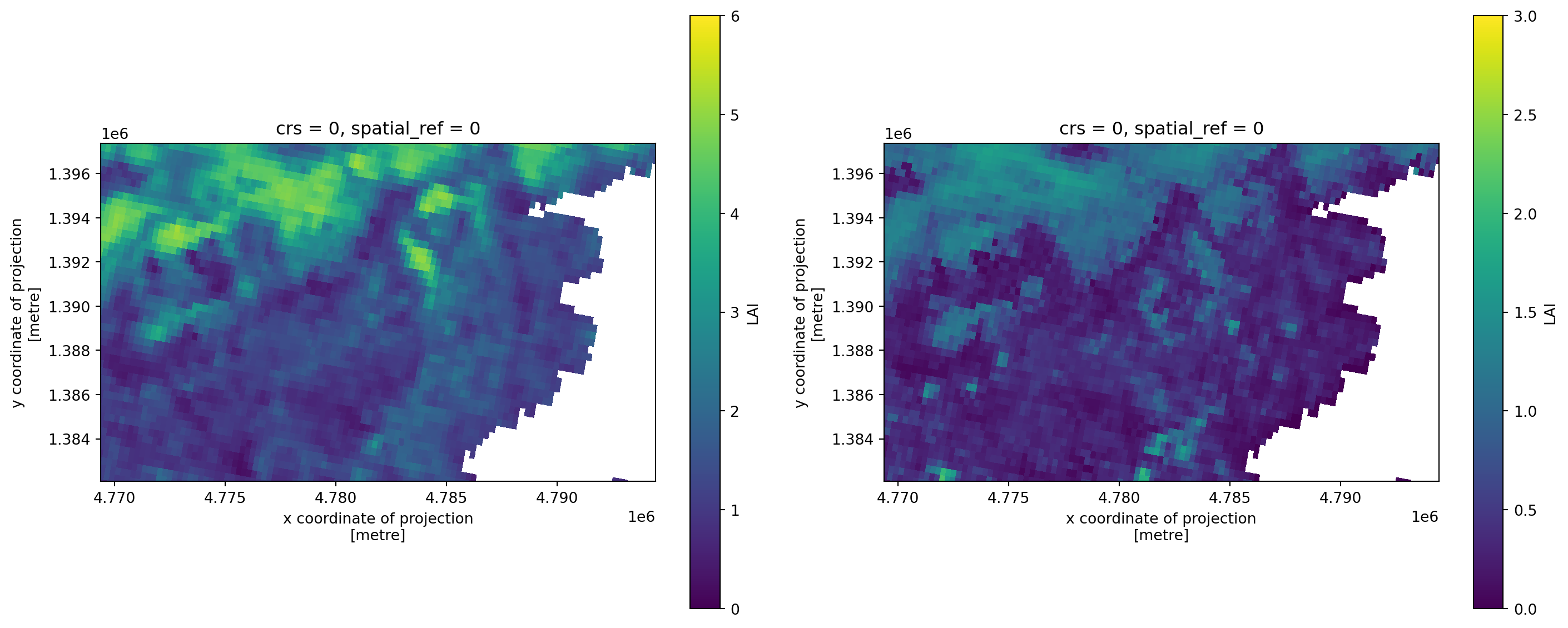
Figure 1: Map of mean LAI (left) and the associated standard deviation (right) for each pixel over time around Lake Garda.
It appears that the northern parts of our study area contain more and variable amounts of green elements per unit area. This might indicate a more complete coverage of foliage and thus forest.
5.2 Timeseries
Now that we have detected possible forested areas, let’s delve a bit deeper into the data. Remember that we deal with a spatiotemporal datacube. This gives us the possibility to study changes for each time increment. Hence we can show what happens to LAI for areas marked with generally low values as well as high values. We can achieve this by filtering the datacube with the where method for areas marked with low and high mean LAI values. In turn we will aggregate the remaining datacube over the spatial dimensions (“x” and “y”) to get a mean values for each time increment.
fig, ax = plt.subplots(1, 2, figsize=(15, 4))
LAI_low = LAI_dc.where(LAI_mean < 4)
LAI_high = LAI_dc.where(LAI_mean > 4)
LAI_low.mean(["x", "y"]).plot.scatter(x="time", ax=ax[0], ylim=(0, 6))
LAI_high.mean(["x", "y"]).plot.scatter(x="time", ax=ax[1], ylim=(0, 6))
ax[0].set_title("Low Mean LAI ($\\bar{LAI} < 4$)")
ax[1].set_title("High Mean LAI ($\\bar{LAI} > 4$)")
plt.tight_layout()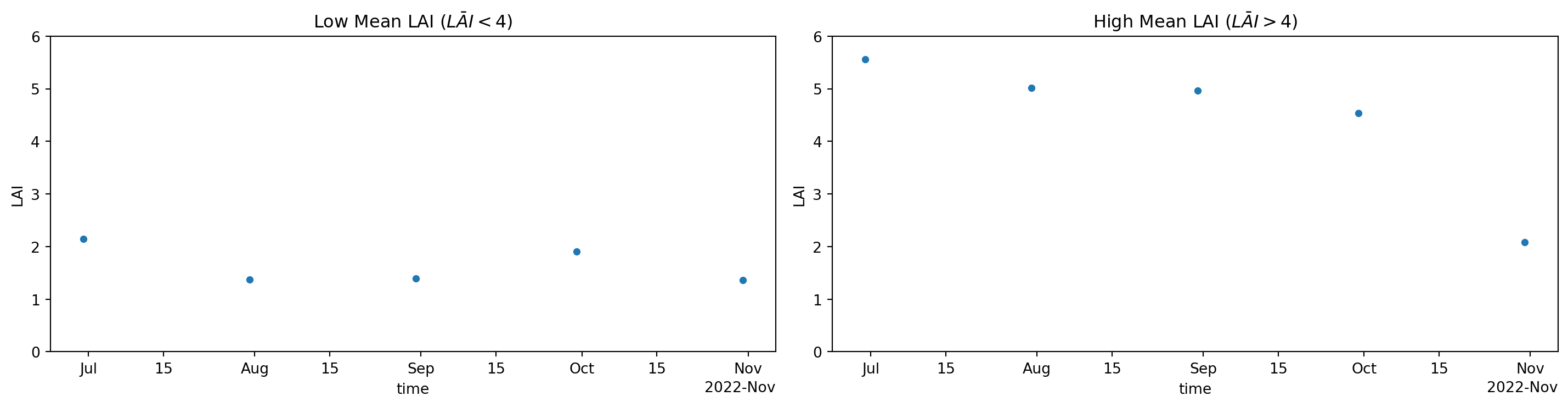
Figure 2: Timeseries of mean LAI per timeslice for areas with low (left) and high (right) mean LAI of Figure1.
Now we can see that areas with high mean LAI values (Figure 1) show a drop-off to values as low as those for areas with low mean LAI during the autumn months (Figure 2 ; right panel). Hence we might deduce that we deal with deciduous forest that becomes less green during autumn, as can be expected for the study area.
Remember that longer wavelengths like L-bands are more likely to penetrate through a forest canopy and would interact more readily with larger object like tree trunks and the forest floor. In turn, C-band microwaves are more likely to interact with sparse and shrub vegetation. The polarization of the emitted and received microwaves is on the other hand dependent on the type of backscattering with co-polarization (HH and VV) happening more frequently with direct backscatter or double bounce scattering. Whereas volume scattering occurs when the radar signal is subject to multiple reflections within 3-dimensional matter, as the orientation of the main scatterers is random, the polarization of the backscattered signal is also random. Volume scattering can therefore cause an increase of cross-polarized intensity.
Let’s put this to the test by checking the microwave backscatter signatures over forested and sparsely vegetated areas as well as water bodies (Lake Garda). Let’s first look at the different sensor readings for the beginning of summer and autumn.
hv.output(widget_location="bottom")
t1 = fused_ds.gam0.\
isel(time=2).\
hvplot.image(robust=True, data_aspect=1, cmap="Greys_r",
rasterize=True, clim=(-25, 0)).\
opts(frame_height=400, aspect="equal")
t2 = fused_ds.gam0.\
isel(time=-1).\
hvplot.image(robust=True, data_aspect=1, cmap="Greys_r",
rasterize=True, clim=(-25, 0)).\
opts(frame_height=400, aspect="equal")
t1 + t2Figure 3: Maps of Sentinel-1 and Alos-2 \(\gamma^0_T \,[dB]\) for the beginning of summer (left) and autumn (right).
The most notable difference is the lower energy received for cross-polarized than for co-polarized microwaves for both Sentinel-1 and Alos-2. The latter differences are independent of the time of year. However, one can also note small changes in the received energy for the same satellite dependent on the time of year. To get a better feel for these changes over time we generate the following interactive plot. On the following plot one can select areas of a certain mean LAI (by clicking on the map) and see the associated timeseries of \(\gamma^0_T\) for each of the sensors.
LAI_image = LAI_mean.hvplot.\
image(rasterize=True, cmap="viridis", clim=(0, 6)).\
opts(title="Mean LAI (Selectable)", frame_height=400, aspect="equal")
def get_timeseries(x, y):
"""
Callback Function Holoviews
Parameters
----------
x: float
numeric value for x selected on LAI map
y: float
numeric value for y selected on LAI map
"""
lai_value = LAI_mean.sel(x=x, y=y, method="nearest").values
if np.isnan(lai_value):
select = fused_ds.where(LAI_mean.isnull())
label = "Water"
else:
mask = np.isclose(LAI_mean, lai_value, atol=0.05)
select = fused_ds.where(mask)
label = "Mean LAI: " + str(np.round(lai_value, 1))
time_series = (
select.gam0.to_dataset("sensor")
.median(["x", "y"], skipna=True)
.hvplot.scatter(ylim=(-30, 5))
.opts(title=label, frame_height=400)
)
return time_series
point_stream = hv.streams.SingleTap(source=LAI_image)
time_series = hv.DynamicMap(get_timeseries, streams=[point_stream])
LAI_image + time_seriesWARNING:param.get_timeseries: Callable raised "ClientResponseError(RequestInfo(url=URL('https://huggingface.co/datasets/martinschobben/microwave-remote-sensing/resolve/main/fused.zarr.zip'), method='GET', headers=<CIMultiDictProxy('Host': 'huggingface.co', 'Range': 'bytes=97824461-103067370', 'Accept': '*/*', 'Accept-Encoding': 'gzip, deflate, br', 'User-Agent': 'Python/3.11 aiohttp/3.11.16')>, real_url=URL('https://huggingface.co/datasets/martinschobben/microwave-remote-sensing/resolve/main/fused.zarr.zip')), (), status=429, message='Too Many Requests', headers=<CIMultiDictProxy('Server': 'CloudFront', 'Date': 'Thu, 10 Apr 2025 13:34:15 GMT', 'Content-Length': '3134', 'Connection': 'keep-alive', 'Content-Type': 'text/html', 'X-Cache': 'Error from cloudfront', 'Via': '1.1 a74378a0e651f6a827eccfaf7700efd2.cloudfront.net (CloudFront)', 'X-Amz-Cf-Pop': 'SFO5-P3', 'X-Amz-Cf-Id': 'KX7b9Tf0VajgyUKE_3zYLbr8DzZ4ImU9EYJ6U8VS8bBLHWVonIcA3w==')>)".
Invoked as get_timeseries(x=None, y=None)--------------------------------------------------------------------------- ClientResponseError Traceback (most recent call last) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/IPython/core/formatters.py:1036, in MimeBundleFormatter.__call__(self, obj, include, exclude) 1033 method = get_real_method(obj, self.print_method) 1035 if method is not None: -> 1036 return method(include=include, exclude=exclude) 1037 return None 1038 else: File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/core/dimension.py:1277, in Dimensioned._repr_mimebundle_(self, include, exclude) 1270 def _repr_mimebundle_(self, include=None, exclude=None): 1271 """ 1272 Resolves the class hierarchy for the class rendering the 1273 object using any display hooks registered on Store.display 1274 hooks. The output of all registered display_hooks is then 1275 combined and returned. 1276 """ -> 1277 return Store.render(self) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/core/options.py:1423, in Store.render(cls, obj) 1421 data, metadata = {}, {} 1422 for hook in hooks: -> 1423 ret = hook(obj) 1424 if ret is None: 1425 continue File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/ipython/display_hooks.py:287, in pprint_display(obj) 285 if not ip.display_formatter.formatters['text/plain'].pprint: 286 return None --> 287 return display(obj, raw_output=True) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/ipython/display_hooks.py:258, in display(obj, raw_output, **kwargs) 256 elif isinstance(obj, (Layout, NdLayout, AdjointLayout)): 257 with option_state(obj): --> 258 output = layout_display(obj) 259 elif isinstance(obj, (HoloMap, DynamicMap)): 260 with option_state(obj): File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/ipython/display_hooks.py:149, in display_hook.<locals>.wrapped(element) 147 try: 148 max_frames = OutputSettings.options['max_frames'] --> 149 mimebundle = fn(element, max_frames=max_frames) 150 if mimebundle is None: 151 return {}, {} File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/ipython/display_hooks.py:223, in layout_display(layout, max_frames) 220 max_frame_warning(max_frames) 221 return None --> 223 return render(layout) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/ipython/display_hooks.py:76, in render(obj, **kwargs) 73 if renderer.fig == 'pdf': 74 renderer = renderer.instance(fig='png') ---> 76 return renderer.components(obj, **kwargs) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/plotting/renderer.py:396, in Renderer.components(self, obj, fmt, comm, **kwargs) 394 embed = (not (dynamic or streams or self.widget_mode == 'live') or config.embed) 395 if embed or config.comms == 'default': --> 396 return self._render_panel(plot, embed, comm) 397 return self._render_ipywidget(plot) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/plotting/renderer.py:403, in Renderer._render_panel(self, plot, embed, comm) 401 doc = Document() 402 with config.set(embed=embed): --> 403 model = plot.layout._render_model(doc, comm) 404 if embed: 405 return render_model(model, comm) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/viewable.py:768, in Viewable._render_model(self, doc, comm) 766 if comm is None: 767 comm = state._comm_manager.get_server_comm() --> 768 model = self.get_root(doc, comm) 770 if self._design and self._design.theme.bokeh_theme: 771 doc.theme = self._design.theme.bokeh_theme File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/layout/base.py:329, in Panel.get_root(self, doc, comm, preprocess) 325 def get_root( 326 self, doc: Document | None = None, comm: Comm | None = None, 327 preprocess: bool = True 328 ) -> Model: --> 329 root = super().get_root(doc, comm, preprocess) 330 # ALERT: Find a better way to handle this 331 if hasattr(root, 'styles') and 'overflow-x' in root.styles: File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/viewable.py:698, in Renderable.get_root(self, doc, comm, preprocess) 696 wrapper = self._design._wrapper(self) 697 if wrapper is self: --> 698 root = self._get_model(doc, comm=comm) 699 if preprocess: 700 self._preprocess(root) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/layout/base.py:313, in Panel._get_model(self, doc, root, parent, comm) 311 root = root or model 312 self._models[root.ref['id']] = (model, parent) --> 313 objects, _ = self._get_objects(model, [], doc, root, comm) 314 props = self._get_properties(doc) 315 props[self._property_mapping['objects']] = objects File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/layout/base.py:295, in Panel._get_objects(self, model, old_objects, doc, root, comm) 293 else: 294 try: --> 295 child = pane._get_model(doc, root, model, comm) 296 except RerenderError as e: 297 if e.layout is not None and e.layout is not self: File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/pane/holoviews.py:437, in HoloViews._get_model(self, doc, root, parent, comm) 435 plot = self.object 436 else: --> 437 plot = self._render(doc, comm, root) 439 plot.pane = self 440 backend = plot.renderer.backend File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/panel/pane/holoviews.py:531, in HoloViews._render(self, doc, comm, root) 528 if comm: 529 kwargs['comm'] = comm --> 531 return renderer.get_plot(self.object, **kwargs) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/plotting/bokeh/renderer.py:68, in BokehRenderer.get_plot(self_or_cls, obj, doc, renderer, **kwargs) 61 @bothmethod 62 def get_plot(self_or_cls, obj, doc=None, renderer=None, **kwargs): 63 """ 64 Given a HoloViews Viewable return a corresponding plot instance. 65 Allows supplying a document attach the plot to, useful when 66 combining the bokeh model with another plot. 67 """ ---> 68 plot = super().get_plot(obj, doc, renderer, **kwargs) 69 if plot.document is None: 70 plot.document = Document() if self_or_cls.notebook_context else curdoc() File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/plotting/renderer.py:216, in Renderer.get_plot(self_or_cls, obj, doc, renderer, comm, **kwargs) 213 raise SkipRendering(msg.format(dims=dims)) 215 # Initialize DynamicMaps with first data item --> 216 initialize_dynamic(obj) 218 if not renderer: 219 renderer = self_or_cls File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/plotting/util.py:277, in initialize_dynamic(obj) 275 continue 276 if not len(dmap): --> 277 dmap[dmap._initial_key()] File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/core/spaces.py:1216, in DynamicMap.__getitem__(self, key) 1214 # Not a cross product and nothing cached so compute element. 1215 if cache is not None: return cache -> 1216 val = self._execute_callback(*tuple_key) 1217 if data_slice: 1218 val = self._dataslice(val, data_slice) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/core/spaces.py:983, in DynamicMap._execute_callback(self, *args) 980 kwargs['_memoization_hash_'] = hash_items 982 with dynamicmap_memoization(self.callback, self.streams): --> 983 retval = self.callback(*args, **kwargs) 984 return self._style(retval) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/holoviews/core/spaces.py:581, in Callable.__call__(self, *args, **kwargs) 578 args, kwargs = (), dict(pos_kwargs, **kwargs) 580 try: --> 581 ret = self.callable(*args, **kwargs) 582 except KeyError: 583 # KeyError is caught separately because it is used to signal 584 # invalid keys on DynamicMap and should not warn 585 raise Cell In[6], line 31, in get_timeseries(x, y) 25 select = fused_ds.where(mask) 26 label = "Mean LAI: " + str(np.round(lai_value, 1)) 28 time_series = ( 29 select.gam0.to_dataset("sensor") 30 .median(["x", "y"], skipna=True) ---> 31 .hvplot.scatter(ylim=(-30, 5)) 32 .opts(title=label, frame_height=400) 33 ) 35 return time_series File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/plotting/core.py:576, in hvPlotTabular.scatter(self, x, y, **kwds) 471 def scatter(self, x=None, y=None, **kwds): 472 """ 473 The `scatter` plot visualizes your points as markers in 2D space. You can visualize 474 one more dimension by using colors. (...) 574 - Wiki: https://en.wikipedia.org/wiki/Scatter_plot 575 """ --> 576 return self(x, y, kind='scatter', **kwds) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/plotting/core.py:95, in hvPlotBase.__call__(self, x, y, kind, **kwds) 92 plot = self._get_converter(x, y, kind, **kwds)(kind, x, y) 93 return pn.panel(plot, **panel_dict) ---> 95 return self._get_converter(x, y, kind, **kwds)(kind, x, y) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/plotting/core.py:102, in hvPlotBase._get_converter(self, x, y, kind, **kwds) 100 y = y or params.pop('y', None) 101 kind = kind or params.pop('kind', None) --> 102 return HoloViewsConverter(self._data, x, y, kind=kind, **params) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/converter.py:626, in HoloViewsConverter.__init__(self, data, x, y, kind, by, use_index, group_label, value_label, backlog, persist, use_dask, crs, fields, groupby, dynamic, grid, legend, rot, title, xlim, ylim, clim, robust, symmetric, logx, logy, loglog, hover, hover_cols, hover_formatters, hover_tooltips, subplots, label, invert, stacked, colorbar, cticks, datashade, rasterize, downsample, resample_when, row, col, debug, framewise, aggregator, projection, global_extent, geo, precompute, flip_xaxis, flip_yaxis, dynspread, x_sampling, y_sampling, pixel_ratio, project, tools, attr_labels, coastline, tiles, tiles_opts, sort_date, check_symmetric_max, transforms, stream, cnorm, features, rescale_discrete_levels, autorange, subcoordinate_y, **kwds) 624 self.value_label = value_label 625 self.label = label --> 626 self._process_data( 627 kind, 628 data, 629 x, 630 y, 631 by, 632 groupby, 633 row, 634 col, 635 use_dask, 636 persist, 637 backlog, 638 label, 639 group_label, 640 value_label, 641 hover_cols, 642 attr_labels, 643 transforms, 644 stream, 645 robust, 646 kwds, 647 ) 649 self.dynamic = dynamic 650 self.geo = any([geo, crs, global_extent, projection, project, coastline, features]) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/converter.py:1160, in HoloViewsConverter._process_data(self, kind, data, x, y, by, groupby, row, col, use_dask, persist, backlog, label, group_label, value_label, hover_cols, attr_labels, transforms, stream, robust, kwds) 1158 other_dims = [] 1159 da = data -> 1160 data, x, y, by_new, groupby_new = process_xarray( 1161 data, 1162 x, 1163 y, 1164 by, 1165 groupby, 1166 use_dask, 1167 persist, 1168 gridded, 1169 label, 1170 value_label, 1171 other_dims, 1172 kind=kind, 1173 ) 1174 if kind == 'rgb' and robust: 1175 # adapted from xarray 1176 # https://github.com/pydata/xarray/blob/6af547cdd9beac3b18420ccb204f801603e11519/xarray/plot/utils.py#L729 1177 vmax = np.nanpercentile(data[z], 100 - 2) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/hvplot/util.py:565, in process_xarray(data, x, y, by, groupby, use_dask, persist, gridded, label, value_label, other_dims, kind) 563 data = data.persist() if persist else data 564 else: --> 565 data = dataset.to_dataframe() 566 if not support_index(data) and len(data.index.names) > 1: 567 data = data.reset_index() File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/dataset.py:7117, in Dataset.to_dataframe(self, dim_order) 7089 """Convert this dataset into a pandas.DataFrame. 7090 7091 Non-index variables in this dataset form the columns of the (...) 7112 7113 """ 7115 ordered_dims = self._normalize_dim_order(dim_order=dim_order) -> 7117 return self._to_dataframe(ordered_dims=ordered_dims) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/dataset.py:7067, in Dataset._to_dataframe(self, ordered_dims) 7057 non_extension_array_columns = [ 7058 k 7059 for k in columns_in_order 7060 if not is_extension_array_dtype(self.variables[k].data) 7061 ] 7062 extension_array_columns = [ 7063 k 7064 for k in columns_in_order 7065 if is_extension_array_dtype(self.variables[k].data) 7066 ] -> 7067 data = [ 7068 self._variables[k].set_dims(ordered_dims).values.reshape(-1) 7069 for k in non_extension_array_columns 7070 ] 7071 index = self.coords.to_index([*ordered_dims]) 7072 broadcasted_df = pd.DataFrame( 7073 dict(zip(non_extension_array_columns, data, strict=True)), index=index 7074 ) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/dataset.py:7068, in <listcomp>(.0) 7057 non_extension_array_columns = [ 7058 k 7059 for k in columns_in_order 7060 if not is_extension_array_dtype(self.variables[k].data) 7061 ] 7062 extension_array_columns = [ 7063 k 7064 for k in columns_in_order 7065 if is_extension_array_dtype(self.variables[k].data) 7066 ] 7067 data = [ -> 7068 self._variables[k].set_dims(ordered_dims).values.reshape(-1) 7069 for k in non_extension_array_columns 7070 ] 7071 index = self.coords.to_index([*ordered_dims]) 7072 broadcasted_df = pd.DataFrame( 7073 dict(zip(non_extension_array_columns, data, strict=True)), index=index 7074 ) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/variable.py:508, in Variable.values(self) 505 @property 506 def values(self) -> np.ndarray: 507 """The variable's data as a numpy.ndarray""" --> 508 return _as_array_or_item(self._data) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/variable.py:302, in _as_array_or_item(data) 288 def _as_array_or_item(data): 289 """Return the given values as a numpy array, or as an individual item if 290 it's a 0d datetime64 or timedelta64 array. 291 (...) 300 TODO: remove this (replace with np.asarray) once these issues are fixed 301 """ --> 302 data = np.asarray(data) 303 if data.ndim == 0: 304 kind = data.dtype.kind File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/dask/array/core.py:1748, in Array.__array__(self, dtype, copy, **kwargs) 1741 if copy is False: 1742 warnings.warn( 1743 "Can't acquire a memory view of a Dask array. " 1744 "This will raise in the future.", 1745 FutureWarning, 1746 ) -> 1748 x = self.compute() 1750 # Apply requested dtype and convert non-numpy backends to numpy. 1751 # If copy is True, numpy is going to perform its own deep copy 1752 # after this method returns. 1753 # If copy is None, finalize() ensures that the returned object 1754 # does not share memory with an object stored in the graph or on a 1755 # process-local Worker. 1756 return np.asarray(x, dtype=dtype) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/dask/base.py:370, in DaskMethodsMixin.compute(self, **kwargs) 346 def compute(self, **kwargs): 347 """Compute this dask collection 348 349 This turns a lazy Dask collection into its in-memory equivalent. (...) 368 dask.compute 369 """ --> 370 (result,) = compute(self, traverse=False, **kwargs) 371 return result File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/dask/base.py:656, in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs) 653 postcomputes.append(x.__dask_postcompute__()) 655 with shorten_traceback(): --> 656 results = schedule(dsk, keys, **kwargs) 658 return repack([f(r, *a) for r, (f, a) in zip(results, postcomputes)]) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/indexing.py:574, in ImplicitToExplicitIndexingAdapter.__array__(self, dtype, copy) 570 def __array__( 571 self, dtype: np.typing.DTypeLike = None, /, *, copy: bool | None = None 572 ) -> np.ndarray: 573 if Version(np.__version__) >= Version("2.0.0"): --> 574 return np.asarray(self.get_duck_array(), dtype=dtype, copy=copy) 575 else: 576 return np.asarray(self.get_duck_array(), dtype=dtype) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/indexing.py:579, in ImplicitToExplicitIndexingAdapter.get_duck_array(self) 578 def get_duck_array(self): --> 579 return self.array.get_duck_array() File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/indexing.py:790, in CopyOnWriteArray.get_duck_array(self) 789 def get_duck_array(self): --> 790 return self.array.get_duck_array() File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/indexing.py:653, in LazilyIndexedArray.get_duck_array(self) 649 array = apply_indexer(self.array, self.key) 650 else: 651 # If the array is not an ExplicitlyIndexedNDArrayMixin, 652 # it may wrap a BackendArray so use its __getitem__ --> 653 array = self.array[self.key] 655 # self.array[self.key] is now a numpy array when 656 # self.array is a BackendArray subclass 657 # and self.key is BasicIndexer((slice(None, None, None),)) 658 # so we need the explicit check for ExplicitlyIndexed 659 if isinstance(array, ExplicitlyIndexed): File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/backends/zarr.py:223, in ZarrArrayWrapper.__getitem__(self, key) 221 elif isinstance(key, indexing.OuterIndexer): 222 method = self._oindex --> 223 return indexing.explicit_indexing_adapter( 224 key, array.shape, indexing.IndexingSupport.VECTORIZED, method 225 ) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/core/indexing.py:1014, in explicit_indexing_adapter(key, shape, indexing_support, raw_indexing_method) 992 """Support explicit indexing by delegating to a raw indexing method. 993 994 Outer and/or vectorized indexers are supported by indexing a second time (...) 1011 Indexing result, in the form of a duck numpy-array. 1012 """ 1013 raw_key, numpy_indices = decompose_indexer(key, shape, indexing_support) -> 1014 result = raw_indexing_method(raw_key.tuple) 1015 if numpy_indices.tuple: 1016 # index the loaded duck array 1017 indexable = as_indexable(result) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/xarray/backends/zarr.py:213, in ZarrArrayWrapper._getitem(self, key) 212 def _getitem(self, key): --> 213 return self._array[key] File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/zarr/core.py:796, in Array.__getitem__(self, selection) 794 result = self.vindex[selection] 795 elif is_pure_orthogonal_indexing(pure_selection, self.ndim): --> 796 result = self.get_orthogonal_selection(pure_selection, fields=fields) 797 else: 798 result = self.get_basic_selection(pure_selection, fields=fields) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/zarr/core.py:1078, in Array.get_orthogonal_selection(self, selection, out, fields) 1075 # setup indexer 1076 indexer = OrthogonalIndexer(selection, self) -> 1078 return self._get_selection(indexer=indexer, out=out, fields=fields) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/zarr/core.py:1341, in Array._get_selection(self, indexer, out, fields) 1338 if math.prod(out_shape) > 0: 1339 # allow storage to get multiple items at once 1340 lchunk_coords, lchunk_selection, lout_selection = zip(*indexer) -> 1341 self._chunk_getitems( 1342 lchunk_coords, 1343 lchunk_selection, 1344 out, 1345 lout_selection, 1346 drop_axes=indexer.drop_axes, 1347 fields=fields, 1348 ) 1349 if out.shape: 1350 return out File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/zarr/core.py:2182, in Array._chunk_getitems(self, lchunk_coords, lchunk_selection, out, lout_selection, drop_axes, fields) 2180 if not isinstance(self._meta_array, np.ndarray): 2181 contexts = ConstantMap(ckeys, constant=Context(meta_array=self._meta_array)) -> 2182 cdatas = self.chunk_store.getitems(ckeys, contexts=contexts) 2184 for ckey, chunk_select, out_select in zip(ckeys, lchunk_selection, lout_selection): 2185 if ckey in cdatas: File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/zarr/storage.py:1426, in FSStore.getitems(self, keys, contexts) 1422 def getitems( 1423 self, keys: Sequence[str], *, contexts: Mapping[str, Context] 1424 ) -> Mapping[str, Any]: 1425 keys_transformed = {self._normalize_key(key): key for key in keys} -> 1426 results_transformed = self.map.getitems(list(keys_transformed), on_error="return") 1427 results = {} 1428 for k, v in results_transformed.items(): File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/mapping.py:105, in FSMap.getitems(self, keys, on_error) 103 oe = on_error if on_error == "raise" else "return" 104 try: --> 105 out = self.fs.cat(keys2, on_error=oe) 106 if isinstance(out, bytes): 107 out = {keys2[0]: out} File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/implementations/cached.py:458, in CachingFileSystem.__getattribute__.<locals>.<lambda>(*args, **kw) 411 def __getattribute__(self, item): 412 if item in { 413 "load_cache", 414 "_open", (...) 456 # all the methods defined in this class. Note `open` here, since 457 # it calls `_open`, but is actually in superclass --> 458 return lambda *args, **kw: getattr(type(self), item).__get__(self)( 459 *args, **kw 460 ) 461 if item in ["__reduce_ex__"]: 462 raise AttributeError File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/implementations/cached.py:651, in WholeFileCacheFileSystem.cat(self, path, recursive, on_error, callback, **kwargs) 648 paths.remove(p) 650 if getpaths: --> 651 self.fs.get(getpaths, storepaths) 652 self.save_cache() 654 callback.set_size(len(paths)) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/spec.py:976, in AbstractFileSystem.get(self, rpath, lpath, recursive, callback, maxdepth, **kwargs) 974 for lpath, rpath in callback.wrap(zip(lpaths, rpaths)): 975 with callback.branched(rpath, lpath) as child: --> 976 self.get_file(rpath, lpath, callback=child, **kwargs) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/spec.py:899, in AbstractFileSystem.get_file(self, rpath, lpath, callback, outfile, **kwargs) 896 fs = LocalFileSystem(auto_mkdir=True) 897 fs.makedirs(fs._parent(lpath), exist_ok=True) --> 899 with self.open(rpath, "rb", **kwargs) as f1: 900 if outfile is None: 901 outfile = open(lpath, "wb") File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/spec.py:1310, in AbstractFileSystem.open(self, path, mode, block_size, cache_options, compression, **kwargs) 1308 else: 1309 ac = kwargs.pop("autocommit", not self._intrans) -> 1310 f = self._open( 1311 path, 1312 mode=mode, 1313 block_size=block_size, 1314 autocommit=ac, 1315 cache_options=cache_options, 1316 **kwargs, 1317 ) 1318 if compression is not None: 1319 from fsspec.compression import compr File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/implementations/zip.py:130, in ZipFileSystem._open(self, path, mode, block_size, autocommit, cache_options, **kwargs) 128 if "r" in self.mode and "w" in mode: 129 raise OSError("ZipFS can only be open for reading or writing, not both") --> 130 out = self.zip.open(path, mode.strip("b"), force_zip64=self.force_zip_64) 131 if "r" in mode: 132 info = self.info(path) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/zipfile.py:1580, in ZipFile.open(self, name, mode, pwd, force_zip64) 1576 zef_file = _SharedFile(self.fp, zinfo.header_offset, 1577 self._fpclose, self._lock, lambda: self._writing) 1578 try: 1579 # Skip the file header: -> 1580 fheader = zef_file.read(sizeFileHeader) 1581 if len(fheader) != sizeFileHeader: 1582 raise BadZipFile("Truncated file header") File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/zipfile.py:786, in _SharedFile.read(self, n) 782 raise ValueError("Can't read from the ZIP file while there " 783 "is an open writing handle on it. " 784 "Close the writing handle before trying to read.") 785 self._file.seek(self._pos) --> 786 data = self._file.read(n) 787 self._pos = self._file.tell() 788 return data File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/implementations/http.py:627, in HTTPFile.read(self, length) 625 else: 626 length = min(self.size - self.loc, length) --> 627 return super().read(length) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/spec.py:2083, in AbstractBufferedFile.read(self, length) 2080 if length == 0: 2081 # don't even bother calling fetch 2082 return b"" -> 2083 out = self.cache._fetch(self.loc, self.loc + length) 2085 logger.debug( 2086 "%s read: %i - %i %s", 2087 self, (...) 2090 self.cache._log_stats(), 2091 ) 2092 self.loc += len(out) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/caching.py:545, in BytesCache._fetch(self, start, end) 543 if self.end is None or self.end - end > self.blocksize: 544 self.total_requested_bytes += bend - start --> 545 self.cache = self.fetcher(start, bend) 546 self.start = start 547 else: File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/asyn.py:118, in sync_wrapper.<locals>.wrapper(*args, **kwargs) 115 @functools.wraps(func) 116 def wrapper(*args, **kwargs): 117 self = obj or args[0] --> 118 return sync(self.loop, func, *args, **kwargs) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/asyn.py:103, in sync(loop, func, timeout, *args, **kwargs) 101 raise FSTimeoutError from return_result 102 elif isinstance(return_result, BaseException): --> 103 raise return_result 104 else: 105 return return_result File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/asyn.py:56, in _runner(event, coro, result, timeout) 54 coro = asyncio.wait_for(coro, timeout=timeout) 55 try: ---> 56 result[0] = await coro 57 except Exception as ex: 58 result[0] = ex File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/fsspec/implementations/http.py:682, in HTTPFile.async_fetch_range(self, start, end) 679 if r.status == 416: 680 # range request outside file 681 return b"" --> 682 r.raise_for_status() 684 # If the server has handled the range request, it should reply 685 # with status 206 (partial content). But we'll guess that a suitable 686 # Content-Range header or a Content-Length no more than the 687 # requested range also mean we have got the desired range. 688 response_is_range = ( 689 r.status == 206 690 or self._parse_content_range(r.headers)[0] == start 691 or int(r.headers.get("Content-Length", end + 1)) <= end - start 692 ) File ~/work/eo-datascience/eo-datascience/.conda_envs/microwave-remote-sensing/lib/python3.11/site-packages/aiohttp/client_reqrep.py:1161, in ClientResponse.raise_for_status(self) 1158 if not self._in_context: 1159 self.release() -> 1161 raise ClientResponseError( 1162 self.request_info, 1163 self.history, 1164 status=self.status, 1165 message=self.reason, 1166 headers=self.headers, 1167 ) ClientResponseError: 429, message='Too Many Requests', url='https://huggingface.co/datasets/martinschobben/microwave-remote-sensing/resolve/main/fused.zarr.zip'
:Layout
.DynamicMap.I :DynamicMap []
:Image [x,y] (LAI)
.DynamicMap.II :DynamicMap []Figure 4: Map of MEAN LAI around Lake Garda. The pixel values can be seen by hovering your mouse over the pixels. Clicking on the pixel will generate the timeseries for the associated mean LAI on the right hand-side. (Right) Timeseries of for Sentinel-1 and Alos-2 \(\gamma^0_T [dB]\).
Can you see some patterns when analyzing the different wavelengths and polarizations?
Remember again that we deal with a logarithmic scale. A measurement of 10 dB is 10 times brighter than the intensity measured at 0 dB, and 100 times brighter at 20 dB. The most notable difference is that the offset between cross- and co-polarised signals becomes larger at low LAI and lower at higher LAI. This might indicate the effect of volume scattering in forested areas where co- and cross-polarization render backscattering values more equal. You will study the differences among cross- and co-polarized backscattering in more detail in the homework exercise.
