import datetime
import holoviews as hv
import hvplot.pandas
import hvplot.xarray
import numpy as np
import pandas as pd
import panel as pn
import pystac_client
import rioxarray # noqa: F401
import xarray as xr
from bokeh.models import FixedTicker
from odc import stac as odc_stac
from scipy.stats import norm
pn.extension()
hv.extension("bokeh")
This notebook contains interactive elements. The full interactive elements can only be viewed on Binder by clicking on the Binder badge or 🚀 button.
B.1 Overview
This notebook explains how microwave (\(\sigma^0\)) backscattering can be used to map the extent of a flood. We replicate in this exercise the work of (Bauer-Marschallinger et al. 2022) on the TU Wien Bayesian-based flood mapping algorithm.
B.2 Prerequisites
| Concepts | Importance | Notes |
|---|---|---|
| Intro to xarray | Necessary | |
| Intro to Harmonic parameters | Necessary | |
| Documentation hvPlot | Helpful | Interactive plotting |
| Documentation odc-stac | Helpful | Data access |
- Time to learn: 10 min
B.3 Imports
B.4 Greece Flooding 2018
In this exercise we will replicate the case study of the above mentioned paper, the February 2018 flooding of the Greek region of Thessaly.
time_range = "2018-02-28T04:00:00Z/2018-02-28T05:00:00Z"
minlon, maxlon = 21.93, 22.23
minlat, maxlat = 39.47, 39.64
bounding_box = [minlon, minlat, maxlon, maxlat]B.5 EODC STAC Catalog
The data required for TU Wien flood mapping algorithm consists of terrain corrected sigma naught backscatter data \(\sigma^{0}\), the projected local incidence angle (PLIA) values of those measurements, and the harmonic parameters (HPAR) of a model fit on the pixel’s backscatter time series. The latter two datasets will needed to calculate the probability density functions over land and water for. We will be getting the required data from the EODC STAC Catalog. Specifically the collections: SENTINEL_SIG0_20M, SENTINEL1_MPLIA and SENTINEL1_HPAR. We use the pystac-client and odc_stac packages to, respectively, discover and fetch the data.
Due to the way the data is acquired and stored, some items include “no data” areas. In our case, no data has the value -9999, but this can vary from data provider to data provider. This information can usually be found in the metadata. Furthermore, to save memory, data is often stored as integer (e.g. 25) and not in float (e.g. 2.5) format. For this reason, the backscatter values are often multiplied by a scale factor. Hence we define the function post_process_eodc_cube to correct for these factors as obtained from the STAC metadata.
B.5.1 Sigma naught
eodc_catalog = pystac_client.Client.open("https://stac.eodc.eu/api/v1")
search = eodc_catalog.search(
collections="SENTINEL1_SIG0_20M",
bbox=bounding_box,
datetime=time_range,
)
items_sig0 = search.item_collection()
def post_process_eodc_cube(dc, items, bands):
"""
Postprocessing of EODC data cubes.
Parameters
----------
x : xarray.Dataset
items: pystac.item_collection.ItemCollection
STAC items that concern the Xarray Dataset
bands: array
Selected bands
Returns
-------
xarray.Dataset
"""
if not isinstance(bands, tuple):
bands = tuple([bands])
for i in bands:
dc[i] = post_process_eodc_cube_(dc[i], items, i)
return dc
def post_process_eodc_cube_(dc, items, band):
fields = items[0].assets[band].extra_fields
scale = fields.get("raster:bands")[0]["scale"]
nodata = fields.get("raster:bands")[0]["nodata"]
return dc.where(dc != nodata) / scale
bands = "VV"
sig0_dc = odc_stac.load(items_sig0, bands=bands, bbox=bounding_box)
sig0_dc = (
post_process_eodc_cube(sig0_dc, items_sig0, bands)
.rename_vars({"VV": "sig0"})
.dropna(dim="time", how="all")
.median("time")
)
sig0_dc<xarray.Dataset> Size: 5MB
Dimensions: (y: 977, x: 1324)
Coordinates:
* y (y) float64 8kB 6.388e+05 6.388e+05 ... 6.193e+05 6.193e+05
* x (x) float64 11kB 5.658e+06 5.658e+06 ... 5.684e+06 5.684e+06
spatial_ref int32 4B 27704
Data variables:
sig0 (y, x) float32 5MB -9.6 -9.2 -8.3 -8.7 ... -12.3 -11.6 -9.7B.5.2 Harmonic Parameters
search = eodc_catalog.search(
collections="SENTINEL1_HPAR",
bbox=bounding_box,
query=["sat:relative_orbit=80"],
)
items_hpar = search.item_collection()
bands = ("C1", "C2", "C3", "M0", "S1", "S2", "S3", "STD")
hpar_dc = odc_stac.load(
items_hpar,
bands=bands,
bbox=bounding_box,
groupby=None,
)
hpar_dc = post_process_eodc_cube(hpar_dc, items_hpar, bands).median("time")
hpar_dc<xarray.Dataset> Size: 41MB
Dimensions: (y: 977, x: 1324)
Coordinates:
* y (y) float64 8kB 6.388e+05 6.388e+05 ... 6.193e+05 6.193e+05
* x (x) float64 11kB 5.658e+06 5.658e+06 ... 5.684e+06 5.684e+06
spatial_ref int32 4B 27704
Data variables:
C1 (y, x) float32 5MB -0.1 -0.1 0.0 0.1 0.3 ... 1.2 1.6 1.8 1.4
C2 (y, x) float32 5MB -0.1 -0.2 -0.2 0.0 -0.1 ... 0.2 0.2 0.6 0.6
C3 (y, x) float32 5MB 0.2 0.1 0.0 0.0 0.1 ... -0.4 -0.6 -0.5 -0.6
M0 (y, x) float32 5MB -9.0 -9.7 -10.0 -9.7 ... -11.8 -11.3 -11.5
S1 (y, x) float32 5MB -0.3 -0.2 -0.2 -0.1 ... -0.3 -0.2 -0.7 -1.1
S2 (y, x) float32 5MB -0.2 0.0 0.0 -0.2 ... -0.2 -0.3 -0.4 -0.2
S3 (y, x) float32 5MB -0.1 0.0 0.0 -0.1 -0.1 ... 0.0 0.1 0.1 0.4
STD (y, x) float32 5MB 1.3 1.2 1.1 1.0 1.2 ... 1.9 1.9 1.8 1.8 1.9B.5.3 Projected Local Incidence Angles
search = eodc_catalog.search(
collections="SENTINEL1_MPLIA",
bbox=bounding_box,
query=["sat:relative_orbit=80"],
)
items_plia = search.item_collection()
bands = "MPLIA"
plia_dc = odc_stac.load(
items_plia,
bands=bands,
bbox=bounding_box,
)
plia_dc = post_process_eodc_cube(plia_dc, items_plia, bands).median("time")
plia_dc<xarray.Dataset> Size: 5MB
Dimensions: (y: 977, x: 1324)
Coordinates:
* y (y) float64 8kB 6.388e+05 6.388e+05 ... 6.193e+05 6.193e+05
* x (x) float64 11kB 5.658e+06 5.658e+06 ... 5.684e+06 5.684e+06
spatial_ref int32 4B 27704
Data variables:
MPLIA (y, x) float32 5MB 27.32 29.22 32.16 ... 33.79 34.02 34.27Finally, we merged the datasets as one big dataset and reproject the data in EPSG 4326 for easier visualizing of the data.
flood_dc = xr.merge([sig0_dc, plia_dc, hpar_dc])
flood_dc = flood_dc.rio.reproject("EPSG:4326").rio.write_crs("EPSG:4326")
flood_dc/tmp/ipykernel_4466/355527453.py:1: FutureWarning: In a future version of xarray the default value for compat will change from compat='no_conflicts' to compat='override'. This is likely to lead to different results when combining overlapping variables with the same name. To opt in to new defaults and get rid of these warnings now use `set_options(use_new_combine_kwarg_defaults=True) or set compat explicitly.
flood_dc = xr.merge([sig0_dc, plia_dc, hpar_dc])<xarray.Dataset> Size: 49MB
Dimensions: (x: 1443, y: 846)
Coordinates:
* x (x) float64 12kB 21.92 21.92 21.92 21.93 ... 22.23 22.23 22.23
* y (y) float64 7kB 39.65 39.65 39.65 39.65 ... 39.46 39.46 39.46
spatial_ref int64 8B 0
Data variables:
sig0 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
MPLIA (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
C1 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
C2 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
C3 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
M0 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
S1 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
S2 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
S3 (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nan
STD (y, x) float32 5MB nan nan nan nan nan ... nan nan nan nan nanB.6 From Backscattering to Flood Mapping
In the following lines we create a map with microwave backscattering values.
mrs_view = flood_dc.sig0.hvplot.image(
x="x", y="y", cmap="viridis", geo=True, tiles=True
).opts(frame_height=400)
mrs_viewB.7 Microwave Backscattering over Land and Water
Reverend Bayes was concerned with two events, one (the hypothesis) occurring before the other (the evidence). If we know its cause, it is easy to logically deduce the probability of an effect. However, in this case we want to deduce the probability of a cause from an observed effect, also known as “reversed probability”. In the case of flood mapping, we have \(\sigma^0\) backscatter observations over land (the effect) and we want to deduce the probability of flooding (\(F\)) and non-flooding (\(NF\)).
In other words, we want to know the probability of flooding \(P(F)\) given a pixel’s \(\sigma^0\):
\[P(F|\sigma^0)\]
and the probability of a pixel being not flooded \(P(NF)\) given a certain \(\sigma^0\):
\[P(NF|\sigma^0).\]
Bayes showed that these can be deduced from the observation that forward and reversed probability are equal, so that:
\[P(F|\sigma^0)P(\sigma^0) = P(\sigma^0|F)P(F)\]
and
\[P(NF|\sigma^0)P(\sigma^0) = P(\sigma^0|NF)P(NF).\]
The forward probability of \(\sigma^0\) given the occurrence of flooding (\(P(\sigma^0|F)\)) and \(\sigma^0\) given no flooding (\(P(\sigma^0|NF)\)) can be extracted from past information on backscattering over land and water surfaces. As seen in the sketch below (Figure B.2), the characteristics of backscattering over land and water differ considerably.
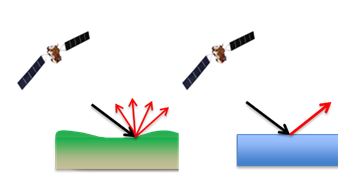
B.8 Likelihoods
The so-called likelihoods of \(P(\sigma^0|F)\) and \(P(\sigma^0|NF)\) can thus be calculated from past backscattering information. In the following code chunk we define the functions calc_water_likelihood and calc_land_likelihood to calculate the water and land likelihood’s of a pixel, based on the Xarray datasets for the PLIA and HPAR, respectively.
def calc_water_likelihood(sigma, x=None, y=None):
"""
Calculate water likelihoods.
Parameters
----------
sigma: float|array
Sigma naught value(s)
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
numpy array
"""
point = flood_dc.sel(x=x, y=y, method="nearest")
wbsc_mean = point.MPLIA * -0.394181 + -4.142015
wbsc_std = 2.754041
return norm.pdf(sigma, wbsc_mean.to_numpy(), wbsc_std)
def expected_land_backscatter(data, dtime_str):
w = np.pi * 2 / 365
dt = datetime.datetime.strptime(dtime_str, "%Y-%m-%d")
t = dt.timetuple().tm_yday
wt = w * t
M0 = data.M0
S1 = data.S1
S2 = data.S2
S3 = data.S3
C1 = data.C1
C2 = data.C2
C3 = data.C3
hm_c1 = (M0 + S1 * np.sin(wt)) + (C1 * np.cos(wt))
hm_c2 = (hm_c1 + S2 * np.sin(2 * wt)) + C2 * np.cos(2 * wt)
hm_c3 = (hm_c2 + S3 * np.sin(3 * wt)) + C3 * np.cos(3 * wt)
return hm_c3
def calc_land_likelihood(sigma, x=None, y=None):
"""
Calculate land likelihoods.
Parameters
----------
sigma: float|array
Sigma naught value(s)
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
numpy array
"""
point = flood_dc.sel(x=x, y=y, method="nearest")
lbsc_mean = expected_land_backscatter(point, "2018-02-01")
lbsc_std = point.STD
return norm.pdf(sigma, lbsc_mean.to_numpy(), lbsc_std.to_numpy())Without going into the details of how these likelihoods are calculated, you can hover over a pixel of the map to plot the likelihoods of \(\sigma^0\) being governed by land or water. For reference we model the water and land likelihoods (model_likelihoods) over a range of \(\sigma^0\) values.
def model_likelihoods(sigma=(-30, 0), x=None, y=None):
"""
Model likelihoods over a range of sigma naught.
Parameters
----------
sigma: tuple
Minimum and maximum for range of sigma naught values
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
Pandas Datafrane
"""
sigma = np.arange(sigma[0], sigma[1], 0.1)
land_likelihood = calc_land_likelihood(sigma=sigma, x=x, y=y)
water_likelihood = calc_water_likelihood(sigma=sigma, x=x, y=y)
point = flood_dc.sel(x=x, y=y, method="nearest")
return pd.DataFrame(
{
"sigma": sigma,
"water_likelihood": water_likelihood,
"land_likelihood": land_likelihood,
"observed": np.repeat(point.sig0.values, len(land_likelihood)),
}
)
pointer = hv.streams.PointerXY(source=mrs_view.get(1), x=22.1, y=39.5)
likelihood_pdi = hvplot.bind(
model_likelihoods, x=pointer.param.x, y=pointer.param.y
).interactive()
view_likelihoods = (
likelihood_pdi.hvplot("sigma", "water_likelihood", ylabel="likelihoods").dmap()
* likelihood_pdi.hvplot("sigma", "land_likelihood").dmap()
* likelihood_pdi.hvplot("observed", "land_likelihood").dmap()
).opts(frame_height=200, frame_width=300)
view_likelihoods + mrs_view.get(1)B.9 Posteriors
Having calculated the likelihoods, we can now move on to calculate the probability of (non-)flooding given a pixel’s \(\sigma^0\). These so-called posteriors need one more piece of information, as can be seen in the equation above. We need the probability that a pixel is flooded \(P(F)\) or not flooded \(P(NF)\). Of course, these are the figures we’ve been trying to find this whole time. We don’t actually have them yet, so what can we do? In Bayesian statistics, we can just start with our best guess. These guesses are called our “priors”, because they are the beliefs we hold prior to looking at the data. This subjective prior belief is the foundation Bayesian statistics, and we use the likelihoods we just calculated to update our belief in this particular hypothesis. This updated belief is called the “posterior”.
Let’s say that our best estimate for the chance of flooding versus non-flooding of a pixel is 50-50: a coin flip. We now can also calculate the probability of backscattering \(P(\sigma^0)\), as the weighted average of the water and land likelihoods, ensuring that our posteriors range between 0 to 1.
The following code block shows how we calculate the priors.
def calc_posteriors(sigma, x=None, y=None):
"""
Calculate posterior probability.
Parameters
----------
sigma: float|array
Sigma naught value(s)
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
Tuple of two Numpy arrays
"""
land_likelihood = calc_land_likelihood(sigma=sigma, x=x, y=y)
water_likelihood = calc_water_likelihood(sigma=sigma, x=x, y=y)
evidence = (water_likelihood * 0.5) + (land_likelihood * 0.5)
water_posterior = (water_likelihood * 0.5) / evidence
land_posterior = (land_likelihood * 0.5) / evidence
return water_posterior, land_posteriorWe can plot the posterior probabilities of flooding and non-flooding again and compare these to pixel’s measured \(\sigma^0\). For reference we model the flood and non-flood posteriors (model_posteriors) over a range of \(\sigma^0\) values. Hover on a pixel to calculate the posterior probability.
def model_posteriors(sigma=(-30, 0), x=None, y=None):
"""
Model posterior probabilities over a range of sigma naught.
Parameters
----------
sigma: tuple
Minimum and maximum for range of sigma naught values
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
Pandas Datafrane
"""
bays_pd = model_likelihoods(sigma=sigma, x=x, y=y)
sigma = np.arange(sigma[0], sigma[1], 0.1)
bays_pd["f_post_prob"], bays_pd["nf_post_prob"] = calc_posteriors(
sigma=sigma, x=x, y=y
)
return bays_pd
posterior_pdi = hvplot.bind(
model_posteriors, x=pointer.param.x, y=pointer.param.y
).interactive()
view_posteriors = (
posterior_pdi.hvplot("sigma", "f_post_prob", ylabel="posteriors").dmap()
* posterior_pdi.hvplot("sigma", "nf_post_prob").dmap()
* posterior_pdi.hvplot("observed", "nf_post_prob").dmap()
).opts(frame_height=200, frame_width=300)
(view_likelihoods + view_posteriors).cols(1) + mrs_view.get(1)B.10 Flood Classification
We are now ready to combine all this information and classify the pixels according to the probability of flooding given the backscatter value of each pixel. Here we just look whether the probability of flooding is higher than non-flooding:
def bayesian_flood_decision(sigma, x=None, y=None):
"""
Bayesian decision.
Parameters
----------
sigma: float|array
Sigma naught value(s)
x: float|array
Longitude
y: float|array
Latitude
Returns
-------
Xarray DataArray
"""
f_post_prob, nf_post_prob = calc_posteriors(sigma=sigma, x=x, y=y)
return xr.where(
np.isnan(f_post_prob) | np.isnan(nf_post_prob),
np.nan,
np.greater(f_post_prob, nf_post_prob),
)Hover on a point in the below map to see the likelihoods and posterior distributions (in the left-hand subplots).
flood_dc["decision"] = (
("y", "x"),
bayesian_flood_decision(flood_dc.sig0, flood_dc.x, flood_dc.y),
)
colorbar_opts = {
"major_label_overrides": {
0: "non-flood",
1: "flood",
},
"ticker": FixedTicker(ticks=[0, 1]),
}
flood_view = flood_dc.decision.hvplot.image(
x="x", y="y", rasterize=True, geo=True, cmap=["rgba(0, 0, 1, 0.1)", "darkred"]
).opts(frame_height=400, colorbar_opts={**colorbar_opts})
mrs_view.get(0) * flood_view