import numpy as np
import intake
import json
import matplotlib.pyplot as plt
import holoviews as hv
from holoviews.streams import RangeXY
from scipy.ndimage import uniform_filter
from functools import partial
hv.extension("bokeh")This notebook will provide an empirical demonstration of speckle - how it originates, how it visually and statistically looks like, and some of the most common approaches to filter it.
Speckle is defined as a kind of noise that affects all radar images. Given the multiple scattering contributions originating from the various elementary objects present within a resolution cell, the resulting backscatter signal can be described as a random constructive and destructive interference of wavelets. As a consequence, speckle is the reason why a granular pattern normally affects SAR images, making it more challenging to interpret and analyze them.
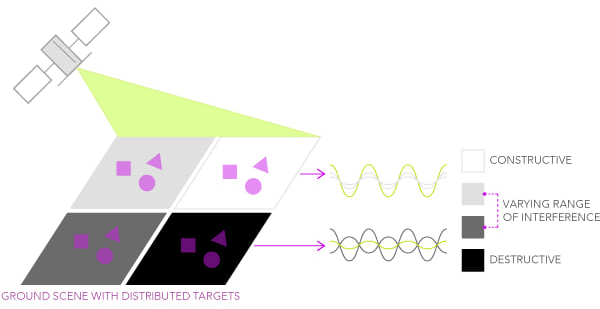
Credits: ESRI
Let’s make an example of a cornfield (with a typical backscattering value of about -10 dB). According to the following equation:
\[ \sigma^0 = \frac{1}{\text{area}} \sum_{n \in \text{area}} \sigma_n \]
We should ideally have a uniform discrete sigma naught \(\sigma^0\) value, given that the cornfield pixel is the only individual contributor.
However, since we already learned from the previous notebooks that a pixel’s ground size can be in the order of tens of meters (i.e., 10 meters for Sentinel-1), we can imagine that different distributed targets in the scene contribute to the global backscattered information.
Let´s replicate this behavior with an ideal uniform area constituted by 100 pixels and then by adding 30% of speckle.
ideal_backscatter = -10 # in dB, a typical value for cornfields
width = 12
size = (width, width)
ideal_data = np.full(size, ideal_backscatter)
ideal_data_linear = 10 ** (
ideal_data / 10
) # Convert dB to linear scale for speckle addition
speckle_fraction = 0.3
num_speckled_pixels = int(
size[0] * size[1] * speckle_fraction
) # Rayleigh speckle noise
speckled_indices = np.random.choice(
width * width, num_speckled_pixels, replace=False
) # random indices for speckle
# Initialize speckled data as the same as the ideal data
speckled_data_linear = ideal_data_linear.copy()
speckle_noise = np.random.gumbel(scale=1.0, size=num_speckled_pixels)
speckled_data_linear.ravel()[speckled_indices] *= (
speckle_noise # Add speckle to the selected pixels
)
ideal_data_dB = 10 * np.log10(ideal_data_linear)
speckled_data_dB = 10 * np.log10(speckled_data_linear)
plt.figure(figsize=(16, 10))
# Ideal data
plt.subplot(2, 2, 1)
plt.imshow(ideal_data_dB, cmap="gray", vmin=-20, vmax=0)
plt.title("Ideal Backscatter (Cornfield)")
plt.colorbar(label="Backscatter (dB)")
# Speckled data
plt.subplot(2, 2, 2)
plt.imshow(speckled_data_dB, cmap="gray", vmin=-20, vmax=0)
plt.title(f"Speckled Backscatter ({int(speckle_fraction * 100)}% of Pixels)")
plt.colorbar(label="Backscatter (dB)")
bins = 25
hist_ideal, bins_ideal = np.histogram(ideal_data_dB.ravel(), bins=bins,
range=(-20, 0))
hist_speckled, bins_speckled = np.histogram(
speckled_data_dB.ravel(), bins=bins, range=(-20, 0)
)
max_freq = max(
hist_ideal.max(), hist_speckled.max()
) # maximum frequency for normalization
# Histogram for ideal data
plt.subplot(2, 2, 3)
plt.hist(ideal_data_dB.ravel(), bins=bins, range=(-20, 0), color="gray",
alpha=0.7)
plt.ylim(0, max_freq)
plt.title("Histogram of Ideal Backscatter")
plt.xlabel("Backscatter (dB)")
plt.ylabel("Frequency")
# Histogram for speckled data
plt.subplot(2, 2, 4)
plt.hist(speckled_data_dB.ravel(), bins=bins, range=(-20, 0), color="gray",
alpha=0.7)
plt.ylim(0, max_freq)
plt.title(
f"Histogram of Speckled Backscatter ({int(speckle_fraction * 100)}%)"
)
plt.xlabel("Backscatter (dB)")
plt.ylabel("Frequency")
plt.tight_layout()/tmp/ipykernel_3338/3970694764.py:26: RuntimeWarning: invalid value encountered in log10
speckled_data_dB = 10 * np.log10(speckled_data_linear)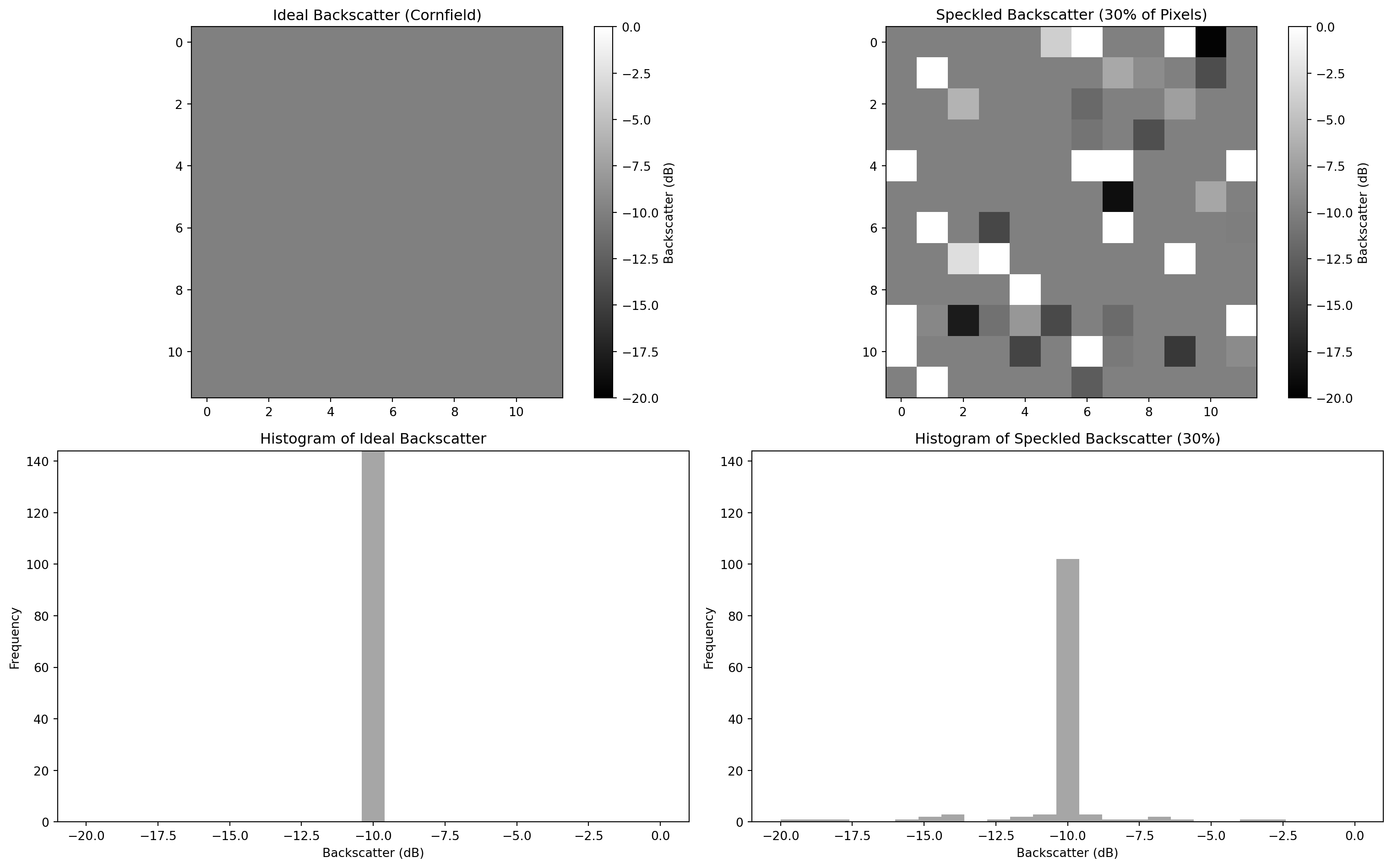
Figure 1: Synthetic data that emulates speckles in microwave backscattering
We can imagine that the second plot represents a real SAR acquisition over a cornfield, while the first plot represents an ideal uniform SAR image over a cornfield land (no speckle). The introduction of a simulated 30% speckle noise could be related to the presence of distributed scatterers of any sort present in the scene, which would cause a pixel-to-pixel variation in terms of intensity.
All the random contributions (such as the wind) would result in a different speckle pattern each time a SAR scene is acquired over the same area. Many subpixel contributors build up a complex scattered pattern in any SAR image, making it erroneous to rely on a single pixel intensity for making reliable image analysis. In order to enhance the degree of usability of a SAR image, several techniques have been put in place to mitigate speckle. We will now show two of the most common approaches: the temporal and the spatial filter.
7.1 Lake Neusiedl data
We load a dataset that contains the CORINE land cover and Sentinel-1 \(\sigma^0_E\) at a 20 meter resolution. This is the same data presented in notebook 6.
url = "https://huggingface.co/datasets/martinschobben/microwave-remote-sensing/resolve/main/microwave-remote-sensing.yml"
cat = intake.open_catalog(url)
fused_ds = cat.speckle.read().compute()
fused_ds<xarray.Dataset> Size: 108MB
Dimensions: (y: 1221, x: 1230, time: 8)
Coordinates:
spatial_ref int64 8B 0
* time (time) datetime64[ns] 64B 2023-08-05T16:51:22 ... 2023-10-28...
* x (x) float64 10kB 5.282e+06 5.282e+06 ... 5.294e+06 5.294e+06
* y (y) float64 10kB 1.571e+06 1.571e+06 ... 1.559e+06 1.559e+06
Data variables:
land_cover (y, x) float64 12MB 12.0 12.0 12.0 12.0 ... 41.0 41.0 41.0 41.0
sig0 (time, y, x) float64 96MB -6.99 -7.32 -8.78 ... -14.32 -14.22We also create the same dashboard for backscatter of different landcover types over time. In order to make this code reusable and adaptable we will define the following function plot_variability, which allows the injection of a spatial and/or temporal filter. It is not important to understand all the code of the following cell!
# Load encoding
with cat.corine_cmap.read()[0] as f:
color_mapping_data = json.load(f)
# Get mapping
color_mapping = {item["value"]: item for item in
color_mapping_data["land_cover"]}
# Get landcover codes present in the image
present_landcover_codes = np.unique(
fused_ds.land_cover.values[~np.isnan(fused_ds.land_cover.values)].
astype(int)
)
def load_image(var_ds, time, land_cover, x_range, y_range,
filter_fun_spatial=None):
"""
Callback Function Landcover.
Parameters
----------
time: panda.datetime
time slice
landcover: int
land cover type
x_range: array_like
longitude range
y_range: array_like
latitude range
Returns
-------
holoviews.Image
"""
if time is not None:
var_ds = var_ds.sel(time=time)
if land_cover == "\xa0\xa0\xa0 Complete Land Cover":
sig0_selected_ds = var_ds.sig0
else:
land_cover_value = int(land_cover.split()[0])
mask_ds = var_ds.land_cover == land_cover_value
sig0_selected_ds = var_ds.sig0.where(mask_ds)
if filter_fun_spatial is not None:
sig0_np = filter_fun_spatial(sig0_selected_ds.values)
else:
sig0_np = sig0_selected_ds.values
# Convert unfiltered data into Holoviews Image
img = hv.Dataset(
(sig0_selected_ds["x"], sig0_selected_ds["y"], sig0_np),
["x", "y"],
"sig0"
)
if x_range and y_range:
img = img.select(x=x_range, y=y_range)
return hv.Image(img)
def plot_variability(var_ds, filter_fun_spatial=None,
filter_fun_temporal=None):
robust_min = var_ds.sig0.quantile(0.02).item()
robust_max = var_ds.sig0.quantile(0.98).item()
bin_edges = [
i + j * 0.5
for i in range(int(robust_min) - 2, int(robust_max) + 2)
for j in range(2)
]
land_cover = {"\xa0\xa0\xa0 Complete Land Cover": 1}
land_cover.update(
{
f"{int(value): 02} {color_mapping[value]['label']}": int(value)
for value in present_landcover_codes
}
)
time = var_ds.sig0["time"].values
rangexy = RangeXY()
if filter_fun_temporal is not None:
var_ds = filter_fun_temporal(var_ds)
load_image_ = partial(
load_image, var_ds=var_ds, filter_fun_spatial=filter_fun_spatial,
time=None
)
dmap = (
hv.DynamicMap(load_image_, kdims=["Landcover"], streams=[rangexy])
.redim.values(Landcover=land_cover)
.hist(normed=True, bins=bin_edges)
)
else:
load_image_ = partial(
load_image, var_ds=var_ds, filter_fun_spatial=filter_fun_spatial
)
dmap = (
hv.DynamicMap(load_image_, kdims=["Time", "Landcover"],
streams=[rangexy])
.redim.values(Time=time, Landcover=land_cover)
.hist(normed=True, bins=bin_edges)
)
image_opts = hv.opts.Image(
cmap="Greys_r",
colorbar=True,
tools=["hover"],
clim=(robust_min, robust_max),
aspect="equal",
framewise=False,
frame_height=500,
frame_width=500,
)
hist_opts = hv.opts.Histogram(width=350, height=555)
return dmap.opts(image_opts, hist_opts)Now, lets work on the real-life dataset to see how speckle actually looks like.
plot_variability(fused_ds)