import urllib.parse
from functools import partial
from pathlib import Path
import folium
import hvplot.xarray # noqa: F401
import numpy as np
import pandas as pd
import rioxarray # noqa: F401
import xarray as xr
from rasterio.enums import Resampling
from rich import print # noqa: A004
from shapely import affinity
from shapely.geometry import box, mapping
from mrs.catalog import make_gitlab_urlsIn this notebook we discuss how we can easily compare images of two or more different time slices, satellites or other earth observation products. We limit our selves to products on a regular grid with an associated coordinate reference system (CRS), known as a raster. This means that each cell of the raster contains an attribute value and location coordinates. The process of combining such rasters to form datacubes is called raster stacking. We can have datacubes in many forms, such as the spatiotemporal datacube:
\[Z = f(x,y,t) \quad \text{,}\]
or when dealing with electromagnetic spectrum, the spectral wavelengths may form an additional dimension of a cube:
\[Z = f(x,y,t, \lambda ) \quad \text{.} \]
We also have already encountered the case where \(Z\) consists of multiple variables, such as seen in the xarray dataset.
\[{Z_1,Z_2,...,Z_3} = f(x,y,t) \]
To perform raster stacking, we generally follow a certain routine (see also Figure 1).
- Collect data (GeoTIFF, NetCDF, Zarr)
- Select an area of interest
- Reproject all rasters to the same projection, resolution, and region
- Stack the individual rasters
To get the same projection, resolution, and region we have to resample one (or more) products. The desired projection, resolution, and region can be adopted from one of the original rasters or it can be a completely new projection of the data.
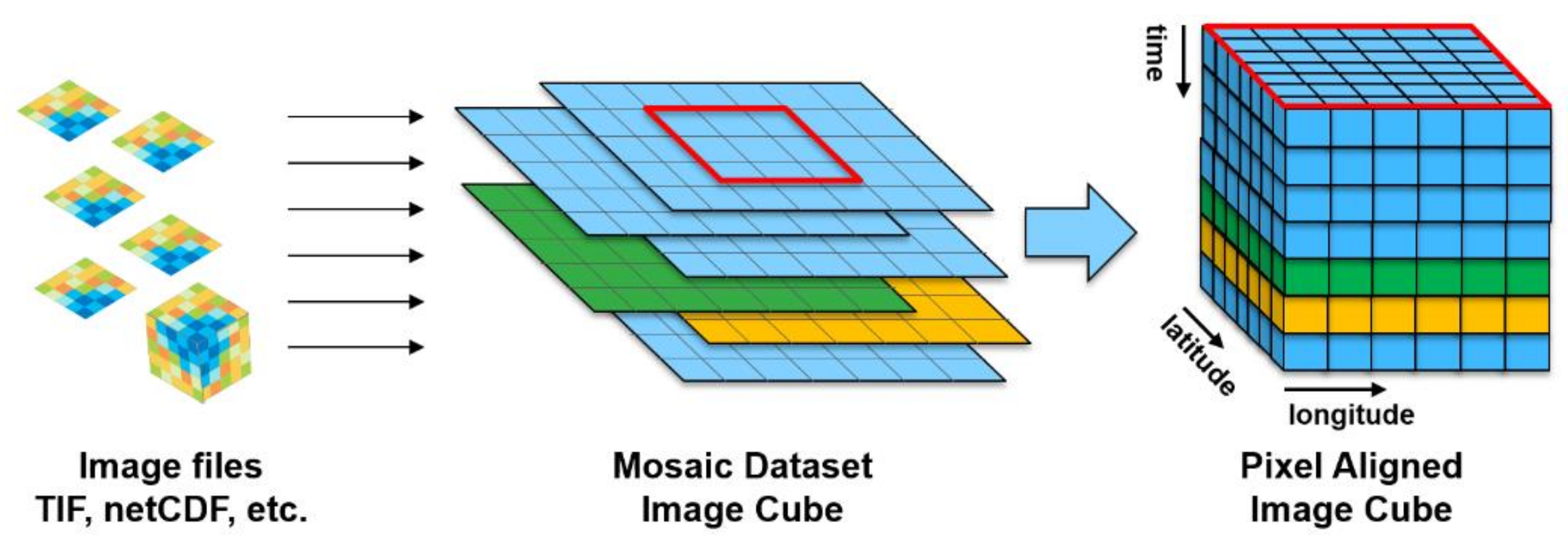
Figure 1: Stacking of arrays to form datacubes (source: https://eox.at).
In this notebook we will study two different SAR products. SAR data from the Advanced Land Observing Satellite (Alos-2), which is a Japanese Satellite platform with an L-band sensor (PALSAR-2) from the Japan Aerospace Exploration Agency (JAXA), and C-band data from the Copernicus Sentinel-1 mission. It is our goal to compare C- with L-band, so we need to somehow stack these arrays.
4.1 Loading Data
Before loading the data into memory we will first look at the area covered by the Sentinel-1 dataset on a map. This way we can select a region of interest for our hypothetical study. We will extract and transform the bounds of the data to longitude and latitude.
bbox = xr.open_mfdataset(
make_gitlab_urls("sentinel-1"),
engine="rasterio",
combine="nested",
concat_dim="band",
).rio.transform_bounds("EPSG:4326")
bbox = box(*bbox)
area_map = folium.Map(
max_bounds=True,
location=[bbox.centroid.y, bbox.centroid.x],
scrollWheelZoom=False,
)
# bounds of image
folium.GeoJson(mapping(bbox), name="Area of Interest", color="red").add_to(area_map)
# minimum longitude, minimum latitude, maximum longitude, maximum latitude
area_of_interest = box(10.3, 45.5, 10.6, 45.6)
folium.GeoJson(mapping(area_of_interest), name="Area of Interest").add_to(area_map)
area_mapFigure 2: Map of study area. Red rectangle is the area covered by the Sentinel-1 raster. Blue rectangle is the area of interest.
On the map we have drawn rectangles of the area covered by the images and of our selected study area. To prevent loading too much data we will now only load the data as defined by the blue rectangle on the folium map.
The Sentinel-1 data is now stored on disk as separate two-dimensional GeoTIFF files with a certain timestamp. The following s1_preprocess function allows to load all files in one go as a spatiotemporal datacube. Basically, the preprocessing function helps reading the timestamp from the file and adds this as a new dimension to the array. The latter allows a concatenation procedure where all files are joined along the new time dimension. In addition by providing area_of_interest.bounds to the parameter bbox we will only load the data of the previously defined area of interest.
def s1_preprocess(
x: xr.Dataset,
bbox: tuple[float, float, float, float],
scale: float,
) -> xr.Dataset:
"""Preprocess file.
Parameters
----------
x : xarray.Dataset
Input raster
bbox: tuple
Minimum longitude minimum latitude maximum longitude maximum latitude
scale: float
Scaling factor
Returns
-------
xarray.Dataset : Output stacked raster
"""
path = Path(urllib.parse.unquote_plus(x.encoding["source"]))
filename = path.parent.name
x = x.rio.clip_box(*bbox, crs="EPSG:4326")
date_str = filename.split("_")[0][1:]
time_str = filename.split("_")[1][:6]
datetime_str = date_str + time_str
date = pd.to_datetime(datetime_str, format="%Y%m%d%H%M%S")
x = x.expand_dims(dim={"time": [date]})
x = (
x.rename({"band_data": "s1_" + path.parent.parent.stem})
.squeeze("band")
.drop_vars("band")
)
return x * scaleWe load the data again with open_mfdataset and by providing the preprocess function, including the bounds of the area of interest and the scaling factor, as follows:
partial_ = partial(s1_preprocess, bbox=area_of_interest.bounds, scale=0.01)
s1_ds = xr.open_mfdataset(
make_gitlab_urls("sentinel-1"),
engine="rasterio",
combine="nested",
chunks=-1,
preprocess=partial_,
compat="no_conflicts",
join="outer",
)4.2 Unlocking Geospatial Information
To enable further stacking of ALOS-2 and Sentinel-1 data we need to know some more information about the raster. Hence we define the following function print_raster to get the projection (CRS), resolution, and region (bounds). The function leverages the functionality of rioxarray; a package for rasters.
def print_raster(raster: xr.Dataset | xr.DataArray, name: str) -> None:
"""Print Raster Metadata.
Parameters
----------
raster: xarray.DataArray|xarray.DataSet
raster to process
name: string
name of product
"""
print(
f"{name} Raster: \n----------------\n"
f"resolution: {raster.rio.resolution()} {raster.rio.crs.units_factor}\n"
f"bounds: {raster.rio.bounds()}\n"
f"CRS: {raster.rio.crs}\n",
)
print_raster(s1_ds, "Sentinel-1")Sentinel-1 Raster: ---------------- resolution: (10.0, -10.0) ('metre', 1.0) bounds: (4769370.0, 1382090.0, 4794450.0, 1397370.0) CRS: EPSG:27704
The CRS “EPSG 27704” is part of the EQUI7Grid. This grid provides equal-area tiles, meaning each tile represents the same area, which helps reducing distorsions. This feature is important for remote sensing as it reduces the so-called oversampling due to geometric distortions when projecting on a sphere. This particular projection is developed by TUWien.
Now we will proceed with loading the ALOS-2 L-band data in much the same fashion as for Sentinel-1. Again timeslices are stored separately as individual GeoTIFFS and they need to be concatenated along the time dimension. We use a slightly different preprocessing function alos_preprocess for this purpose. The most notable difference of this function is the inclusion of a scaling factor for the 16-bit digital numbers (DN):
\[\gamma^0_T = 10 * log_{10}(\text{DN}^2) - 83.0 \,dB\]
to correctly convert the integers to \(\gamma^0_T\) in the dB range.
def alos_preprocess(
x: xr.Dataset,
bbox: tuple[float, float, float, float],
) -> xr.Dataset:
"""Preprocess file.
Parameters
----------
x : xarray.Dataset
Input raster
bbox: tuple
minimum longitude minimum latitude maximum longitude maximum latitude
Returns
-------
xarray.Dataset : Output stacked raster
"""
path = Path(urllib.parse.unquote_plus(x.encoding["source"]))
filename = path.parent.name
x = x.rio.clip_box(*bbox, crs="EPSG:4326")
date_str = filename.split("_")[0][15:22]
date = pd.to_datetime(date_str, format="%y%m%d")
x = x.expand_dims(dim={"time": [date]})
x = (
x.rename({"band_data": "alos_" + path.parent.parent.stem})
.squeeze("band")
.drop_vars("band")
)
# conversion to dB scale of alos
return 10 * np.log10(x**2) - 83.0Now we load the data with the open_mfdataset function of xarray and we provide the preprocessing function (see above), which includes the selection of the bounds of an area of interest and the extraction of time stamps from the file name.
area_of_interest = affinity.scale(area_of_interest, xfact=1.7, yfact=1.7)
partial_ = partial(alos_preprocess, bbox=area_of_interest.bounds)
alos_ds = xr.open_mfdataset(
make_gitlab_urls("alos-2"),
engine="rasterio",
combine="nested",
chunks=-1,
preprocess=partial_,
compat="no_conflicts",
join="outer",
)Also, for this dataset we will look at the metadata in order to compare it with Sentinel-1.
print_raster(alos_ds, "ALOS-2")ALOS-2 Raster: ---------------- resolution: (25.0, -25.0) ('metre', 1.0) bounds: (593137.5, 5035287.5, 633312.5, 5054912.5) CRS: EPSG:32632
4.3 Reprojecting
The ALOS-2 is projected on an UTM grid. We would therefore like to reproject this data to match the projection of Sentinel-1. Furthermore, we will upsample the data to match the Sentinel-1 sampling. The rioxarray package has a very convenient method that can do this all in one go:reproject_match. For continuous data it is best to use a bilinear resampling strategy. As always you have to consider again that we deal with values in the dB range, so we need to convert to the linear scale before bilinear resampling.
alos_ds_lin = 10 ** (alos_ds / 10) # type: ignore[invalid-assignment]
alos_ds_lin = alos_ds_lin.rio.reproject_match( # type: ignore[unresolved-attribute]
s1_ds,
resampling=Resampling.bilinear,
)
alos_ds = 10 * np.log10(alos_ds_lin)We will overwrite the coordinate values of ALOS-2 with those of Sentinel-1. If we would not do this last step, small errors in how the numbers are stored would prevent stacking of the rasters.
alos_ds = alos_ds.assign_coords(
{
"x": s1_ds.x.data,
"y": s1_ds.y.data,
},
)Lastly, we will turn the xarray.DataSet to an xarray.DataArray where a new dimension will constitute the sensor for measurement (satellite + polarization).
s1_da = s1_ds.to_array(dim="sensor")
alos_da = alos_ds.to_array(dim="sensor")
s1_da<xarray.DataArray (sensor: 2, time: 9, y: 1528, x: 2508)> Size: 276MB
dask.array<stack, shape=(2, 9, 1528, 2508), dtype=float32, chunksize=(1, 1, 1528, 2508), chunktype=numpy.ndarray>
Coordinates:
* sensor (sensor) object 16B 's1_VH' 's1_VV'
* time (time) datetime64[us] 72B 2022-06-25T05:27:26 ... 2022-10-11...
* y (y) float64 12kB 1.397e+06 1.397e+06 ... 1.382e+06 1.382e+06
* x (x) float64 20kB 4.769e+06 4.769e+06 ... 4.794e+06 4.794e+06
spatial_ref int64 8B 04.4 Stacking of Multiple Arrays
Now we are finally ready to stack Sentinel-1 C-band and ALOS-2 L-band arrays with the function concat of xarray. Now we can use the newly defined "sensor" dimension to concatenate the two arrays.
fused_da = xr.concat([s1_da, alos_da], dim="sensor", join="outer").rename("gam0")
fused_da<xarray.DataArray 'gam0' (sensor: 4, time: 14, y: 1528, x: 2508)> Size: 858MB
dask.array<concatenate, shape=(4, 14, 1528, 2508), dtype=float32, chunksize=(2, 1, 1094, 1094), chunktype=numpy.ndarray>
Coordinates:
* sensor (sensor) object 32B 's1_VH' 's1_VV' 'alos_HH' 'alos_HV'
* time (time) datetime64[us] 112B 2022-06-25 ... 2022-10-15
* y (y) float64 12kB 1.397e+06 1.397e+06 ... 1.382e+06 1.382e+06
* x (x) float64 20kB 4.769e+06 4.769e+06 ... 4.794e+06 4.794e+06
spatial_ref int64 8B 0The measurements for both satellites don’t occur at the same time. Hence the cube is now padded with 2-D arrays entirely filled with NaN (Not A Number) for some time slices. As we have learned in notebook 2 we can use the resample method to make temporally coherent timeslices for each month. To deal with the dB scale backscatter values as well as the low number of observations per month we use a median of the samples. As taking the median only sorts the samples according to the sample quantiles we do not have to convert the observations to the linear scale.
fused_da = fused_da.resample(time="ME", skipna=True).median().compute()/home/runner/work/microwave-remote-sensing/microwave-remote-sensing/.conda_envs/microwave-remote-sensing/lib/python3.13/site-packages/dask/_task_spec.py:767: RuntimeWarning: All-NaN slice encountered
return self.func(*new_argspec, **kwargs)We can plot each of the variables: “ALOS-2” and “Sentinel-1” to check our results.
fused_da.hvplot.image(robust=True, data_aspect=1, cmap="Greys_r", rasterize=True).opts(
frame_height=600,
aspect="equal",
)Figure 3: Stacked array with ALOS-2 L-band and Sentinel-1 C-band \(\gamma^0_T (dB)\).
