import json
import holoviews as hv
import intake
import numpy as np
import rioxarray # noqa: F401
import xarray as xr
from matplotlib import pyplot as plt
from matplotlib.colors import BoundaryNorm, ListedColormap
from mrs.catalog import CorineColorCollection, get_intake_url
from mrs.plot import plot_corine_data, plot_variability_over_time
hv.extension("bokeh") # type: ignore[too-many-positional-arguments]6.1 Load Sentinel-1 Data
For our analysis we are using sigma naught backscattering data from Sentinel-1. The images that we are analyzing cover the region south of Vienna and west of Lake Neusiedl. We will now load the data and and apply again a preprocessing function. Here we extract the scaling factor and the date the image was taken from the metadata. We will focus our attention to a smaller area containing a part of the Lake Neusiedl Lake and its surrounding land. The obtainedxarray dataset and is then converted to an array, because we only have one variable, the VV backscatter values.
url = get_intake_url()
cat = intake.open_catalog(url)
sig0_da = cat.neusiedler.read().sig0.compute()https://git.geo.tuwien.ac.at/public_projects/microwave-remote-sensing/-/raw/main/microwave-remote-sensing.ymlLet’s have a look at the data by plotting the first timeslice.
sig0_da.isel(time=0).plot(robust=True, cmap="Greys_r").axes.set_aspect("equal")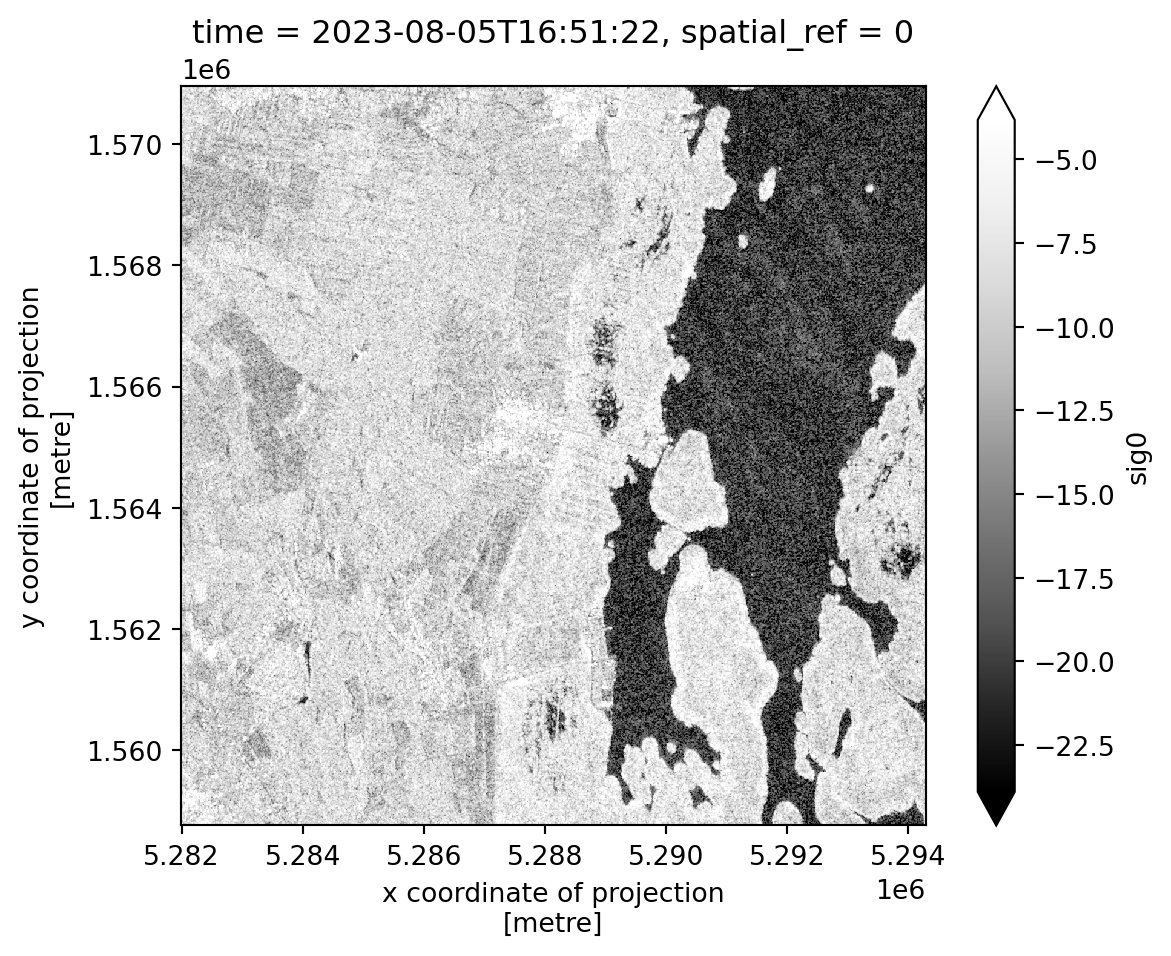
6.2 Load CORINE Landcover Data
We will load the CORINE Land Cover, which is a pan-European land cover and land use inventory with 44 thematic classes. The resolution of this classification is 100 by 100m and the file was created in 2018 (CORINE Land Cover).
cor_da = cat.corine.read().land_cover.compute()6.2.1 Colormapping and Encoding
For the different land cover types we use the official color encoding which can be found in CORINE Land Cover.
# Load encoding and validate with Pydantic
with cat.corine_cmap.read()[0] as f:
corine_colors = CorineColorCollection.model_validate(json.load(f))
# Create cmap and norm for plotting
colors = [info["color"].as_hex() for info in corine_colors.items]
categories = [info["value"] for info in corine_colors.items]
cmap = ListedColormap(colors)
norm = BoundaryNorm([*categories, max(categories) + 1], len(categories))Now we can plot the CORINE Land Cover dataset.
present_landcover_codes = np.unique(cor_da.values[~np.isnan(cor_da.values)].astype(int))
# Create the plot
plot_corine_data(cor_da, cmap, norm, corine_colors, present_landcover_codes)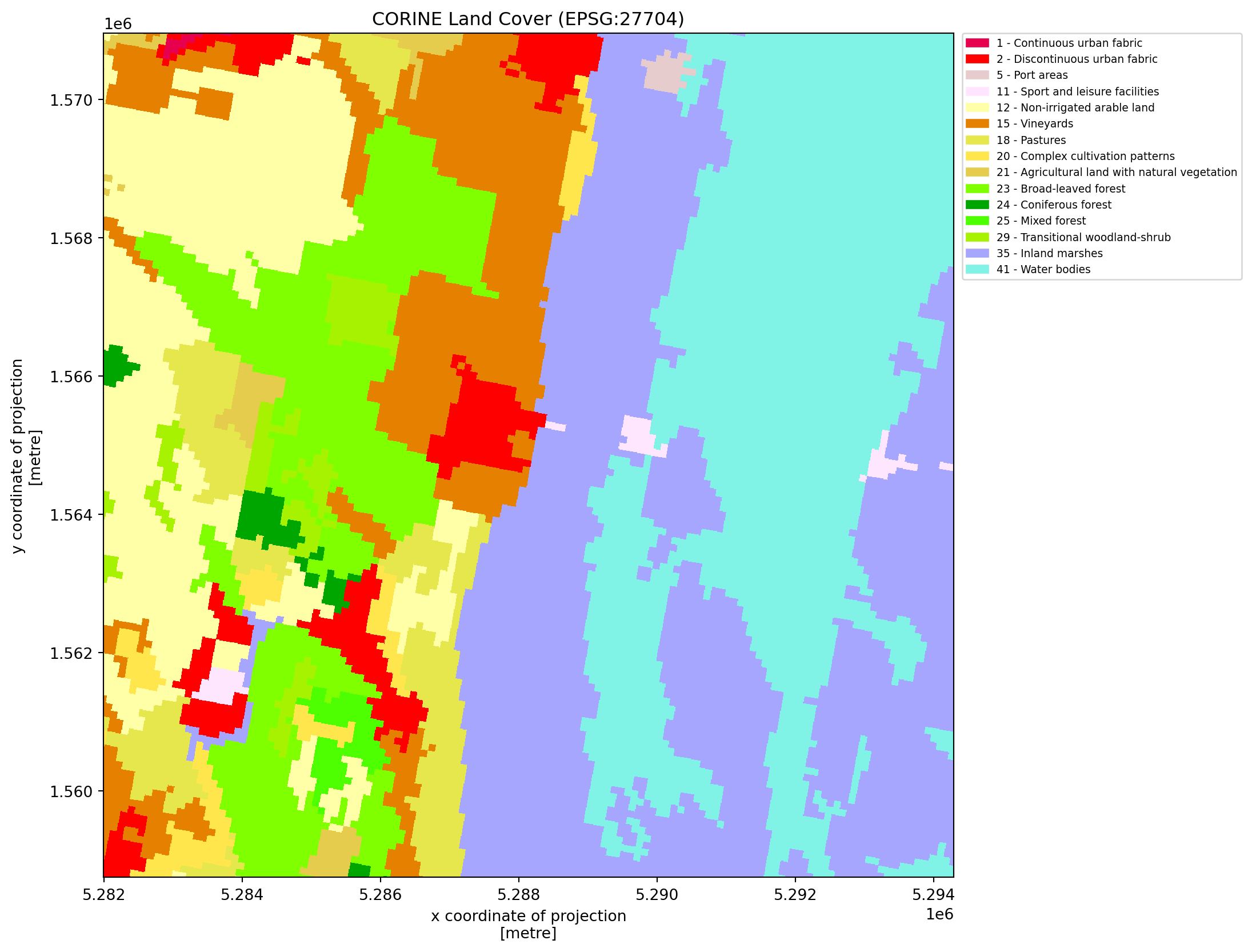
Now we are ready to merge the backscatter data (sig0_da) with the land cover dataset (cor_da) to have one dataset combining all data.
var_ds = xr.merge([sig0_da, cor_da], compat="no_conflicts").drop_vars("band")
var_ds<xarray.Dataset> Size: 96MB
Dimensions: (time: 7, x: 1230, y: 1221)
Coordinates:
* time (time) datetime64[ns] 56B 2023-08-17T16:51:22 ... 2023-10-28...
* x (x) float64 10kB 5.282e+06 5.282e+06 ... 5.294e+06 5.294e+06
* y (y) float64 10kB 1.571e+06 1.571e+06 ... 1.559e+06 1.559e+06
spatial_ref int64 8B 0
Data variables:
sig0 (time, y, x) float64 84MB -8.5 -10.65 -12.07 ... -14.32 -14.22
land_cover (y, x) float64 12MB 12.0 12.0 12.0 12.0 ... 41.0 41.0 41.0 41.06.3 Backscatter Variability
With this combined dataset we can study the backscatter variability in relation to natural media. For example, we can analyze the backscatter variability for water by clipping the dataset to include only the water land cover class, as shown below.
# 41 = encoded value for water bodies
encoded_value_for_waterbodies = 41
waterbodies_mask = var_ds.land_cover == encoded_value_for_waterbodies
waterbodies_mask.plot().axes.set_aspect("equal")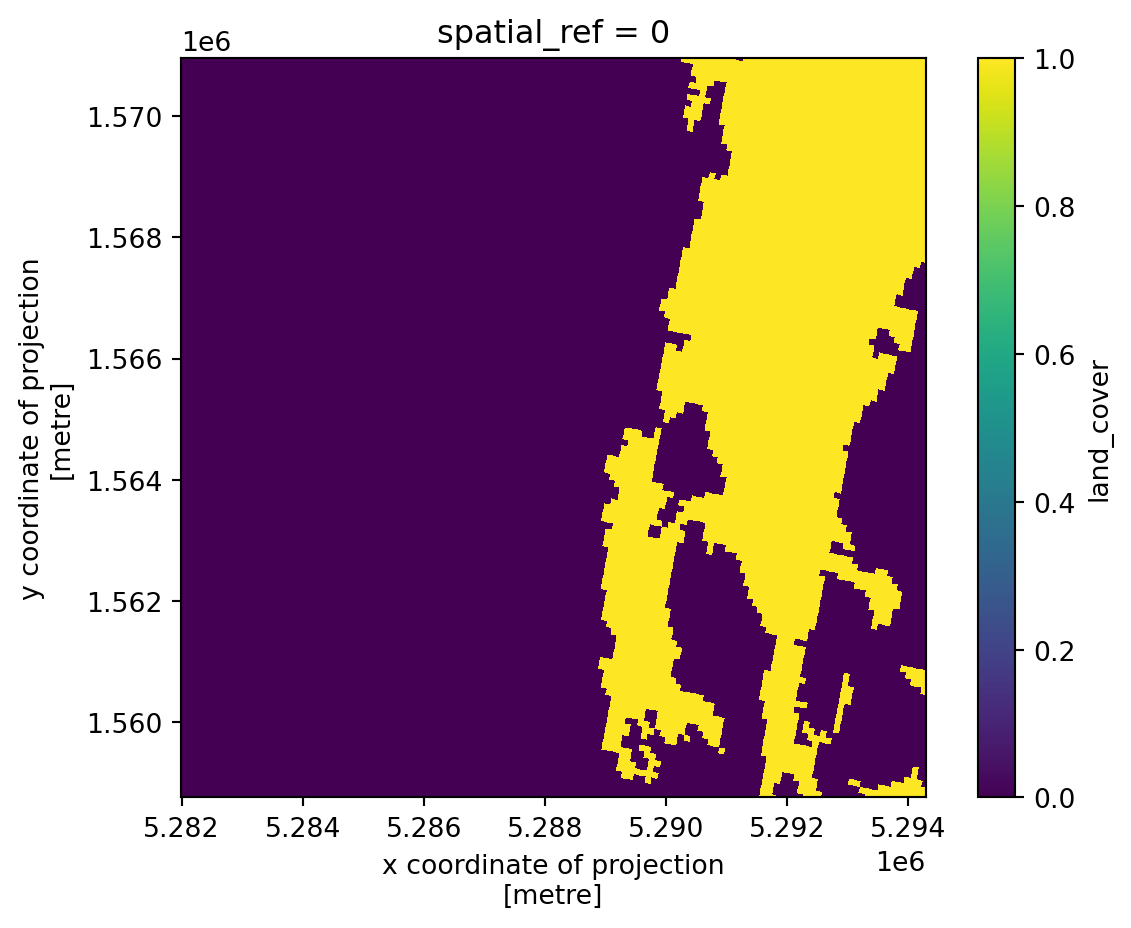
This gives use backscatter values over water only.
waterbodies_sig0 = var_ds.sig0.isel(time=0).where(waterbodies_mask)
waterbodies_sig0.plot(robust=True, cmap="Greys_r").axes.set_aspect("equal")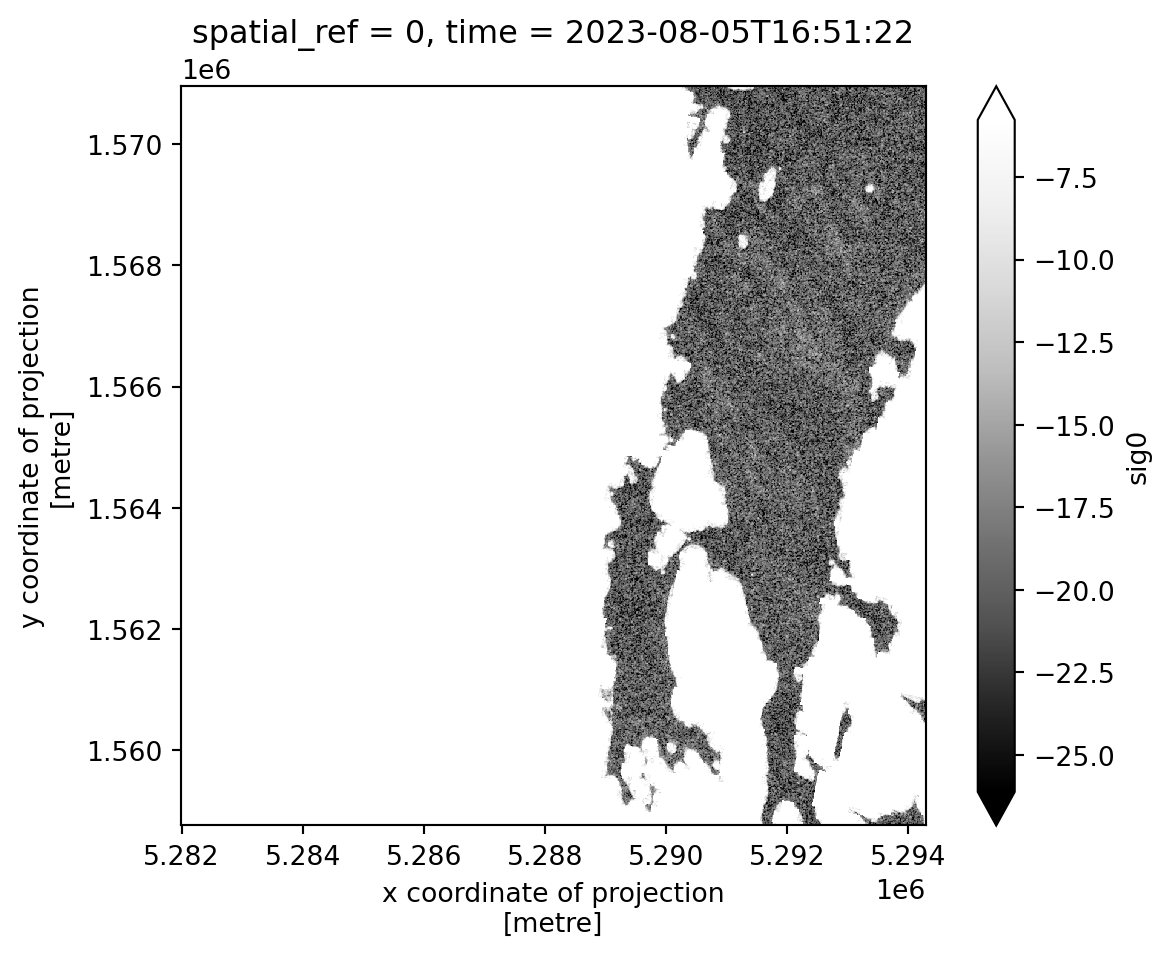
To get an idea of the variability we can create a histogram. Radar backscatter from water bodies fluctuates with surface roughness, which changes with wind conditions, creating spatial and temporal variations in signal intensity.
waterbodies_sig0.plot.hist(bins=50, edgecolor="black")
plt.show()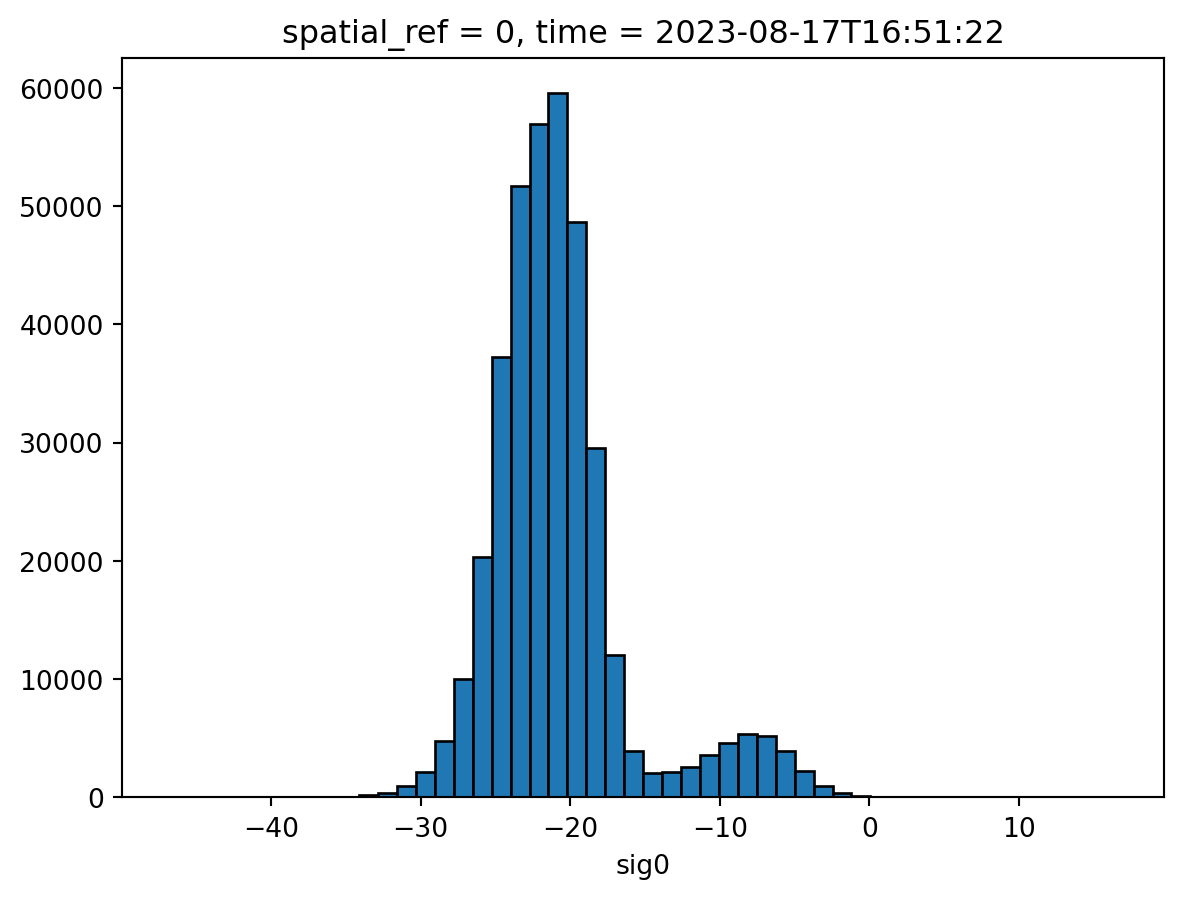
6.4 Variability over Time
Next we will look at the changes in variability in backscatter values over time for each of the CORINE Land Cover types. We do this by creating the following interactive plot.
We can observe that backscatter in agricultural fields varies throughout the year due to several factors:
Seasonal cycles such as planting, growing, and harvesting, which alter the vegetation structure.
Soil moisture variations driven by irrigation or rainfall events.
Crop phenology, where different growth stages affect canopy density and structure.
Canopy moisture dynamics, influencing how radar signals are scattered and absorbed.
plot_variability_over_time(
var_ds=var_ds,
color_mapping=corine_colors.to_dict(),
present_landcover_codes=present_landcover_codes,
)