import json
import holoviews as hv
import intake
import numpy as np
import numpy.typing as npt
from scipy.ndimage import uniform_filter
from mrs.catalog import CorineColorCollection, get_intake_url
from mrs.plot import (
plot_histograms_speckled_and_ideal_data,
plot_variability,
plot_variability_over_time,
)
from mrs.processing import add_speckle, convert_db_to_linear, convert_linear_to_db
hv.extension("bokeh") # type: ignore[too-many-positional-arguments]Let’s make an example of a cornfield (with a typical backscattering value of about -10 dB). According to the following equation:
\[ \sigma^0 = \frac{1}{\text{area}} \sum_{n \in \text{area}} \sigma_n \]
We should ideally have a uniform discrete sigma naught \(\sigma^0\) value, given that the cornfield pixel is the only individual contributor.
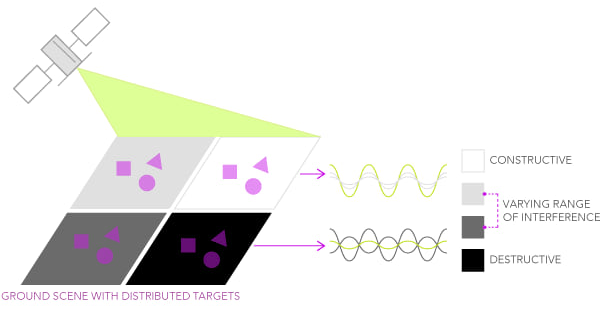
However, since we already learned from the previous notebooks that a pixel’s ground size can be in the order of tens of meters (i.e., 10 meters for Sentinel-1), we can imagine that different distributed targets in the scene contribute to the global backscattered information.
Let´s replicate this behavior with an ideal uniform area constituted by 100 pixels and then by adding 30% of speckle.
ideal_backscatter = -10 # in dB, a typical value for cornfields
size = (12, 12)
ideal_data = np.full(shape=size, fill_value=ideal_backscatter)
speckled_data = np.full(shape=size, fill_value=ideal_backscatter)
# Convert dB to linear scale for speckle addition
ideal_data_linear, speckled_data_linear = convert_db_to_linear(
ideal_data,
speckled_data,
)
# define speckle fraction and add them
speckle_fraction = 0.3
speckled_data_linear = add_speckle(speckled_data_linear, noise=speckle_fraction)
# Convert back to dB scale
ideal_data_db, speckled_data_db = convert_linear_to_db(
ideal_data_linear,
speckled_data_linear,
)/home/runner/work/microwave-remote-sensing/microwave-remote-sensing/.conda_envs/microwave-remote-sensing/lib/python3.13/site-packages/mrs/processing.py:242: RuntimeWarning: invalid value encountered in log10
speckled_data_db = 10 * np.log10(speckled_data_linear)plot_histograms_speckled_and_ideal_data(
backscatter_db_ideal=ideal_data_db,
backscatter_db_speckle=speckled_data_db,
noise=speckle_fraction,
)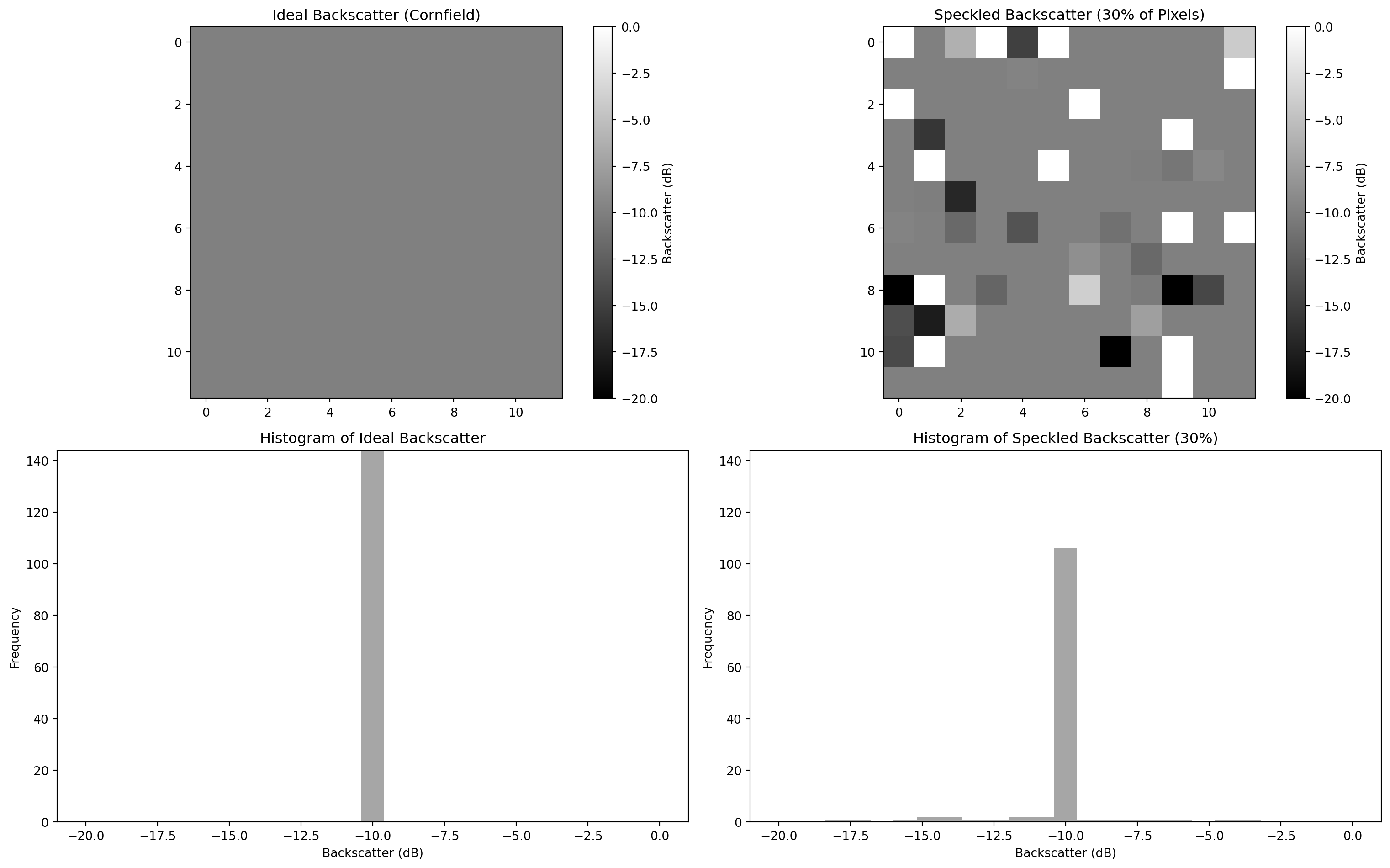
Figure 1: Synthetic data that emulates speckles in microwave backscattering
We can imagine that the second plot represents a real SAR acquisition over a cornfield, while the first plot represents an ideal uniform SAR image over a cornfield land (no speckle). The introduction of a simulated 30% speckle noise could be related to the presence of distributed scatterers of any sort present in the scene, which would cause a pixel-to-pixel variation in terms of intensity.
All the random contributions (such as the wind) would result in a different speckle pattern each time a SAR scene is acquired over the same area. Many subpixel contributors build up a complex scattered pattern in any SAR image, making it erroneous to rely on a single pixel intensity for making reliable image analysis. In order to enhance the degree of usability of a SAR image, several techniques have been put in place to mitigate speckle. We will now show two of the most common approaches: the temporal and the spatial filter.
7.1 Lake Neusiedl data
We load a dataset that contains the CORINE land cover and Sentinel-1 \(\sigma^0_E\) at a 20 meter resolution. This is the same data presented in notebook 6.
url = get_intake_url()
cat = intake.open_catalog(url)
fused_ds = cat.speckle.read().compute()
fused_dshttps://git.geo.tuwien.ac.at/public_projects/microwave-remote-sensing/-/raw/main/microwave-remote-sensing.yml<xarray.Dataset> Size: 96MB
Dimensions: (y: 1221, x: 1230, time: 7)
Coordinates:
* y (y) float64 10kB 1.571e+06 1.571e+06 ... 1.559e+06 1.559e+06
* x (x) float64 10kB 5.282e+06 5.282e+06 ... 5.294e+06 5.294e+06
* time (time) datetime64[ns] 56B 2023-08-17T16:51:22 ... 2023-10-28...
spatial_ref int64 8B 0
Data variables:
land_cover (y, x) float64 12MB 12.0 12.0 12.0 12.0 ... 41.0 41.0 41.0 41.0
sig0 (time, y, x) float64 84MB -8.5 -10.65 -12.07 ... -14.32 -14.22We also create the same dashboard for backscatter of different landcover types over time. In order to make this code reusable and adaptable we will define the following function plot_variability, which allows the injection of a spatial and/or temporal filter. It is not important to understand all the code of the following cell!
# Load encoding and validate with Pydantic
with cat.corine_cmap.read()[0] as f:
corine_colors = CorineColorCollection.model_validate(json.load(f))
# Get mapping
color_mapping = {item["value"]: item for item in corine_colors.items}
# Get landcover codes present in the image
present_landcover_codes = np.unique(
fused_ds.land_cover.where(~np.isnan(fused_ds.land_cover)).astype(int),
)Now, lets work on the real-life dataset to see how speckle actually looks like.
plot_variability_over_time(
var_ds=fused_ds,
color_mapping=corine_colors.to_dict(),
present_landcover_codes=present_landcover_codes,
)Figure 2: Lake Neusiedl \(\sigma^0_E\) without any filter.
The speckle noise typically appears as a “salt-and-pepper” pattern. Also, please note the distribution of backscatter for each land cover. Even though speckle is known for following non-normal distributions (i.e., Rayleigh distribution for amplitude in the linear domain, and the Gumple for intensity in the log domain), we can assume that due to the Central Limit Theorem, the overall backscatter means (dB) tend to follow a Gaussian distribution.
We can mitigate speckle (it is impossible to remove it completely) by following approaches such as: - spatial filtering - taking mean backscatter value over the same land cover, or - temporal filtering - taking the average backscatter value over some time period.
Either way, one pixel is never representative of ground truth! Therefore we need to look at samples and distributions.
7.2 Spatial filtering
We first introduce a common spatial filter. The Lee filter is an adaptive speckle filter. The filter works using a kernel window with a configurable size, which refers to the dimensions of the neighborhood over which the filter operates. The kernel slides across the data, applying the smoothing operation at each pixel position of the image. It follows three assumptions:
- SAR speckle is modeled as a multiplicative noise - the brighter the area the noisier the data.
- The noise and the signal are statistically independent of each other.
- The sample mean and sample variance of a pixel is equal to its local mean and local variance.
This approach comes with some limitations: it reduces the spatial resolution of the SAR image.
Let’s build up a function for applying a Lee filter with a kernel window size of 7 (do not forget to switch back to linear units before doing this computation and to dB after it):
def lee_filter(raster: npt.NDArray, size: int = 7) -> npt.NDArray:
"""Apply the lee-filter to an Image array.
Parameters
----------
raster: ndarray
2D array representing the noisy image (e.g., radar image with speckle)
size: int
Size of the kernel window for the filter (must be odd, default is 7)
Returns
-------
filtered_image (ndarray): The filtered image with reduced speckle noise
"""
raster = np.nan_to_num(raster)
linear = 10 ** (raster / 10)
# Mean and variance over local kernel window
mean_window = uniform_filter(linear, size=size)
mean_sq_window = uniform_filter(linear**2, size=size)
variance_window = mean_sq_window - mean_window**2
# Noise variance estimation (this could also be set manually)
overall_variance = np.var(linear)
# Compute the Lee filter
weights = variance_window / (variance_window + overall_variance)
return 10 * np.log10(mean_window + weights * (linear - mean_window))plot_variability(
var_ds=fused_ds,
color_mapping=color_mapping,
present_landcover_codes=present_landcover_codes,
filter_spatial=lee_filter,
)